[English] 日本語
 Yorodumi
Yorodumi- EMDB-40531: E. coli 50S intermediate, bL17-depletion strain, class: rl17-E-b10 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | E. coli 50S intermediate, bL17-depletion strain, class: rl17-E-b10 | |||||||||
 Map data Map data | rl17-E-b10 Class Particle stack from (doi: 10.1016/j.cell.2016.11.020.) was reanalyzed by CryoSPARC ab-initio subclassification. Aligned to 50S reference and resample to same gird. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribosome / 50S intermediate / RNP | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.45 Å | |||||||||
 Authors Authors | Sheng K / Williamson JR | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Assembly landscape for the bacterial large ribosomal subunit. Authors: Kai Sheng / Ning Li / Jessica N Rabuck-Gibbons / Xiyu Dong / Dmitry Lyumkis / James R Williamson /  Abstract: Assembly of ribosomes in bacteria is highly efficient, taking ~2-3 min, but this makes the abundance of assembly intermediates very low, which is a challenge for mechanistic understanding. Genetic ...Assembly of ribosomes in bacteria is highly efficient, taking ~2-3 min, but this makes the abundance of assembly intermediates very low, which is a challenge for mechanistic understanding. Genetic perturbations of the assembly process create bottlenecks where intermediates accumulate, facilitating structural characterization. We use cryo-electron microscopy, with iterative subclassification to identify intermediates in the assembly of the 50S ribosomal subunit from E. coli. The analysis of the ensemble of intermediates that spans the entire biogenesis pathway for the 50 S subunit was facilitated by a dimensionality reduction and cluster picking approach using PCA-UMAP-HDBSCAN. The identity of the cooperative folding units in the RNA with associated proteins is revealed, and the hierarchy of these units reveals a complete assembly map for all RNA and protein components. The assembly generally proceeds co-transcriptionally, with some flexibility in the landscape to ensure efficiency for this central cellular process under a variety of growth conditions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40531.map.gz emd_40531.map.gz | 11.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40531-v30.xml emd-40531-v30.xml emd-40531.xml emd-40531.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40531_fsc.xml emd_40531_fsc.xml | 5.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_40531.png emd_40531.png | 98 KB | ||
| Others |  emd_40531_half_map_1.map.gz emd_40531_half_map_1.map.gz emd_40531_half_map_2.map.gz emd_40531_half_map_2.map.gz | 14.5 MB 14.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40531 http://ftp.pdbj.org/pub/emdb/structures/EMD-40531 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40531 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40531 | HTTPS FTP |
-Validation report
| Summary document |  emd_40531_validation.pdf.gz emd_40531_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40531_full_validation.pdf.gz emd_40531_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_40531_validation.xml.gz emd_40531_validation.xml.gz | 12.7 KB | Display | |
| Data in CIF |  emd_40531_validation.cif.gz emd_40531_validation.cif.gz | 15.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40531 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40531 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40531 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40531 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40531.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40531.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | rl17-E-b10 Class Particle stack from (doi: 10.1016/j.cell.2016.11.020.) was reanalyzed by CryoSPARC ab-initio subclassification. Aligned to 50S reference and resample to same gird. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.62 Å | ||||||||||||||||||||||||||||||||||||
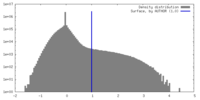
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: rl17-E-b10 Class Particle stack from (doi: 10.1016/j.cell.2016.11.020.) was...
| File | emd_40531_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | rl17-E-b10 Class Particle stack from (doi: 10.1016/j.cell.2016.11.020.) was reanalyzed by CryoSPARC ab-initio subclassification. | ||||||||||||
| Projections & Slices |
| ||||||||||||
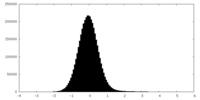
| Density Histograms |
-Half map: rl17-E-b10 Class Particle stack from (doi: 10.1016/j.cell.2016.11.020.) was...
| File | emd_40531_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | rl17-E-b10 Class Particle stack from (doi: 10.1016/j.cell.2016.11.020.) was reanalyzed by CryoSPARC ab-initio subclassification. | ||||||||||||
| Projections & Slices |
| ||||||||||||
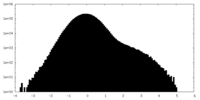
| Density Histograms |
- Sample components
Sample components
-Entire : E. coli 50S assembly intermediate from bL17-depletion strain
| Entire | Name: E. coli 50S assembly intermediate from bL17-depletion strain |
|---|---|
| Components |
|
-Supramolecule #1: E. coli 50S assembly intermediate from bL17-depletion strain
| Supramolecule | Name: E. coli 50S assembly intermediate from bL17-depletion strain type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 33.0 e/Å2 Details: Data were collected with tilts ranging from 0 to 60 degrees at 10-degree increments. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



































































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)