[English] 日本語
 Yorodumi
Yorodumi- EMDB-40504: Cryo-EM structure of TRPM7 N1098Q mutant in GDN detergent in comp... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of TRPM7 N1098Q mutant in GDN detergent in complex with inhibitor NS8593 in closed state | ||||||||||||||||||
 Map data Map data | Cryo-EM structure of TRPM7 N1098Q mutant in GDN detergent in complex with inhibitor NS8593 in closed state | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | transient receptor potential M family member 7 / TRP / channel / TRPM7 / TRP channels / membrane protein / magnesium channel / inhibitor / ligand / NS8593 | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcalcium-dependent cell-matrix adhesion / intracellular magnesium ion homeostasis / zinc ion transport / magnesium ion transmembrane transport / zinc ion transmembrane transporter activity / magnesium ion transmembrane transporter activity / TRP channels / actomyosin structure organization / myosin binding / necroptotic process ...calcium-dependent cell-matrix adhesion / intracellular magnesium ion homeostasis / zinc ion transport / magnesium ion transmembrane transport / zinc ion transmembrane transporter activity / magnesium ion transmembrane transporter activity / TRP channels / actomyosin structure organization / myosin binding / necroptotic process / ruffle / cytoplasmic vesicle membrane / calcium channel activity / calcium ion transport / kinase activity / actin binding / protein autophosphorylation / cytoplasmic vesicle / protein homotetramerization / eukaryotic translation initiation factor 2alpha kinase activity / 3-phosphoinositide-dependent protein kinase activity / DNA-dependent protein kinase activity / ribosomal protein S6 kinase activity / histone H3S10 kinase activity / histone H2AXS139 kinase activity / histone H3S28 kinase activity / histone H4S1 kinase activity / histone H2BS14 kinase activity / histone H3T3 kinase activity / histone H2AS121 kinase activity / Rho-dependent protein serine/threonine kinase activity / histone H2BS36 kinase activity / histone H3S57 kinase activity / histone H2AT120 kinase activity / AMP-activated protein kinase activity / histone H2AS1 kinase activity / histone H3T6 kinase activity / histone H3T11 kinase activity / histone H3T45 kinase activity / non-specific serine/threonine protein kinase / protein kinase activity / protein serine kinase activity / protein serine/threonine kinase activity / ATP binding / metal ion binding / nucleus / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.91 Å | ||||||||||||||||||
 Authors Authors | Nadezhdin KD / Neuberger A / Sobolevsky AI | ||||||||||||||||||
| Funding support |  United States, United States,  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural mechanisms of TRPM7 activation and inhibition. Authors: Kirill D Nadezhdin / Leonor Correia / Chamali Narangoda / Dhilon S Patel / Arthur Neuberger / Thomas Gudermann / Maria G Kurnikova / Vladimir Chubanov / Alexander I Sobolevsky /   Abstract: The transient receptor potential channel TRPM7 is a master regulator of the organismal balance of divalent cations that plays an essential role in embryonic development, immune responses, cell ...The transient receptor potential channel TRPM7 is a master regulator of the organismal balance of divalent cations that plays an essential role in embryonic development, immune responses, cell mobility, proliferation, and differentiation. TRPM7 is implicated in neuronal and cardiovascular disorders, tumor progression and has emerged as a new drug target. Here we use cryo-EM, functional analysis, and molecular dynamics simulations to uncover two distinct structural mechanisms of TRPM7 activation by a gain-of-function mutation and by the agonist naltriben, which show different conformational dynamics and domain involvement. We identify a binding site for highly potent and selective inhibitors and show that they act by stabilizing the TRPM7 closed state. The discovered structural mechanisms provide foundations for understanding the molecular basis of TRPM7 channelopathies and drug development. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40504.map.gz emd_40504.map.gz | 96.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40504-v30.xml emd-40504-v30.xml emd-40504.xml emd-40504.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40504.png emd_40504.png | 82.7 KB | ||
| Filedesc metadata |  emd-40504.cif.gz emd-40504.cif.gz | 7.3 KB | ||
| Others |  emd_40504_half_map_1.map.gz emd_40504_half_map_1.map.gz emd_40504_half_map_2.map.gz emd_40504_half_map_2.map.gz | 95 MB 95 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40504 http://ftp.pdbj.org/pub/emdb/structures/EMD-40504 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40504 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40504 | HTTPS FTP |
-Related structure data
| Related structure data |  8siaMC  8si2C  8si3C  8si4C  8si5C  8si6C  8si7C  8si8C  8sibC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40504.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40504.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of TRPM7 N1098Q mutant in GDN detergent in complex with inhibitor NS8593 in closed state | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_40504_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
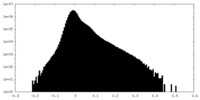
| Density Histograms |
-Half map: Half Map 2
| File | emd_40504_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
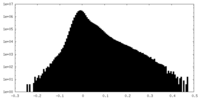
| Density Histograms |
- Sample components
Sample components
-Entire : sample 1
| Entire | Name: sample 1 |
|---|---|
| Components |
|
-Supramolecule #1: sample 1
| Supramolecule | Name: sample 1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 700 KDa |
-Macromolecule #1: Transient receptor potential cation channel subfamily M member 7
| Macromolecule | Name: Transient receptor potential cation channel subfamily M member 7 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 146.902891 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SQKSWIESTL TKRECVYIIP SSKDPHRCLP GCQICQQLVR CFCGRLVKQH ACFTASLAMK YSDVKLGEHF NQAIEEWSVE KHTEQSPTD AYGVINFQGG SHSYRAKYVR LSYDTKPEII LQLLLKEWQM ELPKLVISVH GGMQKFELHP RIKQLLGKGL I KAAVTTGA ...String: SQKSWIESTL TKRECVYIIP SSKDPHRCLP GCQICQQLVR CFCGRLVKQH ACFTASLAMK YSDVKLGEHF NQAIEEWSVE KHTEQSPTD AYGVINFQGG SHSYRAKYVR LSYDTKPEII LQLLLKEWQM ELPKLVISVH GGMQKFELHP RIKQLLGKGL I KAAVTTGA WILTGGVNTG VAKHVGDALK EHASRSSRKI CTIGIAPWGV IENRNDLVGR DVVAPYQTLL NPLSKLNVLN NL HSHFILV DDGTVGKYGA EVRLRRELEK TINQQRIHAR IGQGVPVVAL IFEGGPNVIL TVLEYLQESP PVPVVVCEGT GRA ADLLAY IHKQTEEGGN LPDAAEPDII STIKKTFNFG QSEAVHLFQT MMECMKKKEL ITVFHIGSED HQDIDVAILT ALLK GTNAS AFDQLILTLA WDRVDIAKNH VFVYGQQWLV GSLEQAMLDA LVMDRVSFVK LLIENGVSMH KFLTIPRLEE LYNTK QGPT NPMLFHLIRD VKQGNLPPGY KITLIDIGLV IEYLMGGTYR CTYTRKRFRL IYNSLGGNNR RSGRNTSSST PQLRKS HET FGNRADKKEK MRHNHFIKTA QPYRPKMDAS MEEGKKKRTK DEIVDIDDPE TKRFPYPLNE LLIWACLMKR QVMARFL WQ HGEESMAKAL VACKIYRSMA YEAKQSDLVD DTSEELKQYS NDFGQLAVEL LEQSFRQDET MAMKLLTYEL KNWSNSTC L KLAVSSRLRP FVAHTCTQML LSDMWMGRLN MRKNSWYKVI LSILVPPAIL MLEYKTKAEM SHIPQSQDAH QMTMEDSEN NFHNITEEIP MEVFKEVKIL DSSDGKNEME IHIKSKKLPI TRKFYAFYHA PIVKFWFNTL AYLGFLMLYT FVVLVKMEQL PSVQEWIVI AYIFTYAIEK VREVFMSEAG KISQKIKVWF SDYFNVSDTI AIISFFVGFG LRFGAKWNYI NAYDNHVFVA G RLIYCLNI IFWYVRLLDF LAVNQQAGPY VMMIGKMVAN MFYIVVIMAL VLLSFGVPRK AILYPHEEPS WSLAKDIVFH PY WMIFGEV YAYEIDVCAN DSTLPTICGP GTWLTPFLQA VYLFVQYIIM VNLLIAFFNQ VYLQVKAISN IVWKYQRYHF IMA YHEKPV LPPPLIILSH IVSLFCCVCK RRKKDKTSDG PKLFLTEEDQ KKLHDFEEQC VEMYFDEKDD KFNSGSEERI RVTF ERVEQ MSIQIKEVGD RVNYIKRSLQ SLDSQIGHLQ DLSALTVDTL KTLTAQKASE ASKVHNEITR ELSISKHLAQ NLID UniProtKB: Transient receptor potential cation channel subfamily M member 7 |
-Macromolecule #2: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 2 / Number of copies: 52 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Macromolecule #3: 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13...
| Macromolecule | Name: 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol type: ligand / ID: 3 / Number of copies: 4 / Formula: DU0 |
|---|---|
| Molecular weight | Theoretical: 516.752 Da |
| Chemical component information |  ChemComp-DU0: |
-Macromolecule #4: N-[(1R)-1,2,3,4-tetrahydronaphthalen-1-yl]-1H-benzimidazol-2-amine
| Macromolecule | Name: N-[(1R)-1,2,3,4-tetrahydronaphthalen-1-yl]-1H-benzimidazol-2-amine type: ligand / ID: 4 / Number of copies: 4 / Formula: 6RA |
|---|---|
| Molecular weight | Theoretical: 263.337 Da |
| Chemical component information |  ChemComp-6RA: |
-Macromolecule #5: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #6: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 6 / Number of copies: 4 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||||
| Grid | Model: C-flat-1.2/1.3 / Support film - Material: GOLD / Support film - topology: HOLEY | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||||||||
| Details | mouse TRPM7 |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 11520 pixel / Digitization - Dimensions - Height: 8184 pixel / Number grids imaged: 1 / Number real images: 7659 / Average exposure time: 2.6 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-8sia: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)