[English] 日本語
 Yorodumi
Yorodumi- EMDB-4033: In situ atomic-resolution structure of the baseplate antenna comp... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4033 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ atomic-resolution structure of the baseplate antenna complex in Chlorobaculum tepidum obtained combining solid-state NMR spectroscopy, cryo electron microscopy and polarization spectroscopy | |||||||||||||||
 Map data Map data | 3D reconstruction of the carotenosome CsmA-Bchl a baseplate obtained by cryo electron microscopy | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | photosynthesis / light-harvesting protein / binds bacteriochlorophyll a / oligomeric complex / bacteriochlorophyll binding protein | |||||||||||||||
| Function / homology | chlorosome envelope / Bacteriochlorophyll c-binding protein / Bacteriochlorophyll c-binding superfamily / Bacteriochlorophyll C binding protein / bacteriochlorophyll binding / photosynthesis / metal ion binding / Bacteriochlorophyll c-binding protein Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Chlorobaculum tepidum (bacteria) / Chlorobaculum tepidum (bacteria) /  Chlorobium tepidum (strain ATCC 49652 / DSM 12025 / NBRC 103806 / TLS) (bacteria) Chlorobium tepidum (strain ATCC 49652 / DSM 12025 / NBRC 103806 / TLS) (bacteria) | |||||||||||||||
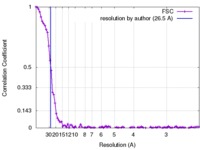
| Method | single particle reconstruction / cryo EM / Resolution: 26.5 Å | |||||||||||||||
 Authors Authors | Nielsen JT / Kulminskaya NV | |||||||||||||||
| Funding support |  Denmark, Denmark,  Estonia, Estonia,  Hungary, 4 items Hungary, 4 items
| |||||||||||||||
 Citation Citation | Journal: Angew Chem Int Ed Engl / Year: 2012 Title: In situ solid-state NMR spectroscopy of protein in heterogeneous membranes: the baseplate antenna complex of Chlorobaculum tepidum. Authors: Natalia V Kulminskaya / Marie Ø Pedersen / Morten Bjerring / Jarl Underhaug / Mette Miller / Niels-Ulrik Frigaard / Jakob T Nielsen / Niels Chr Nielsen /  Abstract: A clever combination: an in situ solid-state NMR analysis of CsmA proteins in the heterogeneous environment of the photoreceptor of Chlorobaculum tepidum is reported. Using different combinations of ...A clever combination: an in situ solid-state NMR analysis of CsmA proteins in the heterogeneous environment of the photoreceptor of Chlorobaculum tepidum is reported. Using different combinations of 2D and 3D solid-state NMR spectra, 90 % of the CsmA resonances are assigned and provide on the basis of chemical shift data information about the structure and conformation of CsmA in the CsmA-bacteriochlorophyll a complex. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4033.map.gz emd_4033.map.gz | 10.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4033-v30.xml emd-4033-v30.xml emd-4033.xml emd-4033.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4033_fsc.xml emd_4033_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_4033.png emd_4033.png | 102.4 KB | ||
| Filedesc metadata |  emd-4033.cif.gz emd-4033.cif.gz | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4033 http://ftp.pdbj.org/pub/emdb/structures/EMD-4033 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4033 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4033 | HTTPS FTP |
-Related structure data
| Related structure data |  5lcbMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4033.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4033.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D reconstruction of the carotenosome CsmA-Bchl a baseplate obtained by cryo electron microscopy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Carotenosome organelle consisting of csma proteins and bchla ante...
| Entire | Name: Carotenosome organelle consisting of csma proteins and bchla antenna molecules. |
|---|---|
| Components |
|
-Supramolecule #1: Carotenosome organelle consisting of csma proteins and bchla ante...
| Supramolecule | Name: Carotenosome organelle consisting of csma proteins and bchla antenna molecules. type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Cryo-EM data of the ice-embedded isolated carotenosomes reveals molecular complex structure. Captured images clearly show supramolecular organization of the protein rows, originating from ...Details: Cryo-EM data of the ice-embedded isolated carotenosomes reveals molecular complex structure. Captured images clearly show supramolecular organization of the protein rows, originating from the BChl a - CsmA baseplate |
|---|---|
| Source (natural) | Organism:  Chlorobaculum tepidum (bacteria) / Organelle: carotenosome Chlorobaculum tepidum (bacteria) / Organelle: carotenosome |
-Macromolecule #1: Bacteriochlorophyll c-binding protein
| Macromolecule | Name: Bacteriochlorophyll c-binding protein / type: protein_or_peptide / ID: 1 / Number of copies: 14 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chlorobium tepidum (strain ATCC 49652 / DSM 12025 / NBRC 103806 / TLS) (bacteria) Chlorobium tepidum (strain ATCC 49652 / DSM 12025 / NBRC 103806 / TLS) (bacteria) |
| Molecular weight | Theoretical: 6.160034 KDa |
| Recombinant expression | Organism:  Chlorobaculum tepidum (bacteria) Chlorobaculum tepidum (bacteria) |
| Sequence | String: MSGGGVFTDI LAAAGRIFEV MVEGHWETVG MLFDSLGKGT MRINRNAYGS MGGGSLRGS UniProtKB: Bacteriochlorophyll c-binding protein |
-Macromolecule #2: BACTERIOCHLOROPHYLL A
| Macromolecule | Name: BACTERIOCHLOROPHYLL A / type: ligand / ID: 2 / Number of copies: 14 / Formula: BCL |
|---|---|
| Molecular weight | Theoretical: 911.504 Da |
| Chemical component information |  ChemComp-BCL: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R2/4 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 270 / Average electron dose: 8.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 4.0 µm / Calibrated defocus min: 1.7 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)