+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of FtAlkB | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Enzyme / MEMBRANE PROTEIN | |||||||||
| Function / homology | Fatty acid desaturase domain / Alkane/xylene monooxygenase / Fatty acid desaturase / lipid metabolic process / monooxygenase activity / metal ion binding / plasma membrane / Alkane 1-monooxygenase Function and homology information Function and homology information | |||||||||
| Biological species |  Fontimonas thermophila (bacteria) / synthetic construct (others) Fontimonas thermophila (bacteria) / synthetic construct (others) | |||||||||
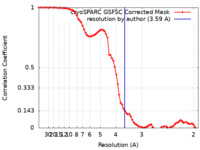
| Method | single particle reconstruction / cryo EM / Resolution: 3.59 Å | |||||||||
 Authors Authors | Zhang J / Feng L | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure and mechanism of the alkane-oxidizing enzyme AlkB. Authors: Xue Guo / Jianxiu Zhang / Lei Han / Juliet Lee / Shoshana C Williams / Allison Forsberg / Yan Xu / Rachel Narehood Austin / Liang Feng /  Abstract: Alkanes are the most energy-rich form of carbon and are widely dispersed in the environment. Their transformation by microbes represents a key step in the global carbon cycle. Alkane monooxygenase ...Alkanes are the most energy-rich form of carbon and are widely dispersed in the environment. Their transformation by microbes represents a key step in the global carbon cycle. Alkane monooxygenase (AlkB), a membrane-spanning metalloenzyme, converts straight chain alkanes to alcohols in the first step of the microbially-mediated degradation of alkanes, thereby playing a critical role in the global cycling of carbon and the bioremediation of oil. AlkB biodiversity is attributed to its ability to oxidize alkanes of various chain lengths, while individual AlkBs target a relatively narrow range. Mechanisms of substrate selectivity and catalytic activity remain elusive. Here we report the cryo-EM structure of AlkB, which provides a distinct architecture for membrane enzymes. Our structure and functional studies reveal an unexpected diiron center configuration and identify molecular determinants for substrate selectivity. These findings provide insight into the catalytic mechanism of AlkB and shed light on its function in alkane-degrading microorganisms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40303.map.gz emd_40303.map.gz | 22.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40303-v30.xml emd-40303-v30.xml emd-40303.xml emd-40303.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40303_fsc.xml emd_40303_fsc.xml | 7.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_40303.png emd_40303.png | 134.3 KB | ||
| Masks |  emd_40303_msk_1.map emd_40303_msk_1.map | 42.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-40303.cif.gz emd-40303.cif.gz | 6.3 KB | ||
| Others |  emd_40303_additional_1.map.gz emd_40303_additional_1.map.gz emd_40303_half_map_1.map.gz emd_40303_half_map_1.map.gz emd_40303_half_map_2.map.gz emd_40303_half_map_2.map.gz | 37.9 MB 39.8 MB 39.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40303 http://ftp.pdbj.org/pub/emdb/structures/EMD-40303 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40303 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40303 | HTTPS FTP |
-Related structure data
| Related structure data |  8sbbMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40303.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40303.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.95 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_40303_msk_1.map emd_40303_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_40303_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_40303_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_40303_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : FtAklB-Nanobody
| Entire | Name: FtAklB-Nanobody |
|---|---|
| Components |
|
-Supramolecule #1: FtAklB-Nanobody
| Supramolecule | Name: FtAklB-Nanobody / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Fontimonas thermophila (bacteria) Fontimonas thermophila (bacteria) |
-Macromolecule #1: Alkane 1-monooxygenase
| Macromolecule | Name: Alkane 1-monooxygenase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Fontimonas thermophila (bacteria) Fontimonas thermophila (bacteria) |
| Molecular weight | Theoretical: 45.56948 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MATSMGSHPG FGADSSGTIA YRDRKRPFWT LSVLWPVSPL IGIYLAHTTG VGAFFWLTLA VWYLIIPLLD WVLGDDQSNP PEAVVPALE SDRYYRILTY LTVPIHYVVL IGSAWYVSTH YGSMTWYDIL GLALSVGIVN GLAINTGHEL GHKKTELERW L AKIVLAVV ...String: MATSMGSHPG FGADSSGTIA YRDRKRPFWT LSVLWPVSPL IGIYLAHTTG VGAFFWLTLA VWYLIIPLLD WVLGDDQSNP PEAVVPALE SDRYYRILTY LTVPIHYVVL IGSAWYVSTH YGSMTWYDIL GLALSVGIVN GLAINTGHEL GHKKTELERW L AKIVLAVV GYGHFFIEHN KGHHKDVATP EDPASAPFGQ SIYRFALREI PGAIIRAWNS EKERLGRLGH SPWSLQNEVL QP LLITIPL YVGLIAVLGV KIIPFLLIQI VFGWWQLTSA NYIEHYGLLR QKLPDGRYER CQPYHSWNSN HVMSNLILFH LQR HSDHHA HPTRRYQSLR DFPDLPTLPS GYPLMFALSY FPPLWRAVMD RRVLDVCGRD ARKINIDPNQ REKIIHKFNL LTG UniProtKB: Alkane 1-monooxygenase |
-Macromolecule #2: Nanobody
| Macromolecule | Name: Nanobody / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 14.502039 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAQVQLQESG GGLVQAGGSL RLSCAASGTI SRYWTMGWYR QAPGKERELV AGISEGGSTN YADSVKGRFT ISRDNAKNTV YLQMNSLKP EDTAVYYCAV TYRGPWFNRD PHYYWGQGTQ VTVSSHHHHH H |
-Macromolecule #3: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Macromolecule #4: DODECANE
| Macromolecule | Name: DODECANE / type: ligand / ID: 4 / Number of copies: 1 / Formula: D12 |
|---|---|
| Molecular weight | Theoretical: 170.335 Da |
| Chemical component information |  ChemComp-D12: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)