[English] 日本語
 Yorodumi
Yorodumi- EMDB-40251: CRISPR-Cas type III-D effector complex bound to self-target RNA i... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
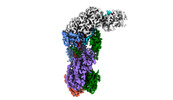
| Title | CRISPR-Cas type III-D effector complex bound to self-target RNA in a post-cleavage state | |||||||||
 Map data Map data | CRISPR-Cas type III-D complex in a post-RNA cleavage state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR / CRISPR-Cas / type III / complex / RNA / crRNA / RNA BINDING PROTEIN-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
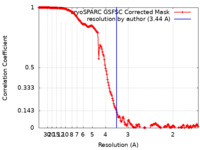
| Method | single particle reconstruction / cryo EM / Resolution: 3.44 Å | |||||||||
 Authors Authors | Schwartz EA / Taylor DW | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: RNA targeting and cleavage by the type III-Dv CRISPR effector complex. Authors: Evan A Schwartz / Jack P K Bravo / Mohd Ahsan / Luis A Macias / Caitlyn L McCafferty / Tyler L Dangerfield / Jada N Walker / Jennifer S Brodbelt / Giulia Palermo / Peter C Fineran / Robert D ...Authors: Evan A Schwartz / Jack P K Bravo / Mohd Ahsan / Luis A Macias / Caitlyn L McCafferty / Tyler L Dangerfield / Jada N Walker / Jennifer S Brodbelt / Giulia Palermo / Peter C Fineran / Robert D Fagerlund / David W Taylor /   Abstract: CRISPR-Cas are adaptive immune systems in bacteria and archaea that utilize CRISPR RNA-guided surveillance complexes to target complementary RNA or DNA for destruction. Target RNA cleavage at regular ...CRISPR-Cas are adaptive immune systems in bacteria and archaea that utilize CRISPR RNA-guided surveillance complexes to target complementary RNA or DNA for destruction. Target RNA cleavage at regular intervals is characteristic of type III effector complexes. Here, we determine the structures of the Synechocystis type III-Dv complex, an apparent evolutionary intermediate from multi-protein to single-protein type III effectors, in pre- and post-cleavage states. The structures show how multi-subunit fusion proteins in the effector are tethered together in an unusual arrangement to assemble into an active and programmable RNA endonuclease and how the effector utilizes a distinct mechanism for target RNA seeding from other type III effectors. Using structural, biochemical, and quantum/classical molecular dynamics simulation, we study the structure and dynamics of the three catalytic sites, where a 2'-OH of the ribose on the target RNA acts as a nucleophile for in line self-cleavage of the upstream scissile phosphate. Strikingly, the arrangement at the catalytic residues of most type III complexes resembles the active site of ribozymes, including the hammerhead, pistol, and Varkud satellite ribozymes. Our work provides detailed molecular insight into the mechanisms of RNA targeting and cleavage by an important intermediate in the evolution of type III effector complexes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40251.map.gz emd_40251.map.gz | 483.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40251-v30.xml emd-40251-v30.xml emd-40251.xml emd-40251.xml | 23.8 KB 23.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40251_fsc.xml emd_40251_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_40251.png emd_40251.png | 44.2 KB | ||
| Filedesc metadata |  emd-40251.cif.gz emd-40251.cif.gz | 7.9 KB | ||
| Others |  emd_40251_half_map_1.map.gz emd_40251_half_map_1.map.gz emd_40251_half_map_2.map.gz emd_40251_half_map_2.map.gz | 475.3 MB 475.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40251 http://ftp.pdbj.org/pub/emdb/structures/EMD-40251 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40251 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40251 | HTTPS FTP |
-Validation report
| Summary document |  emd_40251_validation.pdf.gz emd_40251_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40251_full_validation.pdf.gz emd_40251_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_40251_validation.xml.gz emd_40251_validation.xml.gz | 26.6 KB | Display | |
| Data in CIF |  emd_40251_validation.cif.gz emd_40251_validation.cif.gz | 34.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40251 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40251 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40251 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40251 | HTTPS FTP |
-Related structure data
| Related structure data |  8s9xMC  8s9tC  8s9uC  8s9vC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40251.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40251.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CRISPR-Cas type III-D complex in a post-RNA cleavage state | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8332 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map B
| File | emd_40251_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
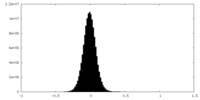
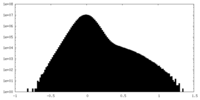
| Density Histograms |
-Half map: half map A
| File | emd_40251_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
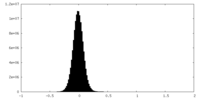
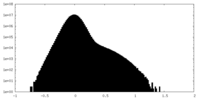
| Density Histograms |
- Sample components
Sample components
-Entire : CRISPR-Cas type III-D effector complex
| Entire | Name: CRISPR-Cas type III-D effector complex |
|---|---|
| Components |
|
-Supramolecule #1: CRISPR-Cas type III-D effector complex
| Supramolecule | Name: CRISPR-Cas type III-D effector complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 332 KDa |
-Macromolecule #1: Cas7-Cas5-Cas11
| Macromolecule | Name: Cas7-Cas5-Cas11 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 87.558938 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRGIEITITM QSDWHVGTGM GRGELDSVVQ RDGDNLPYIP GKTLTGILRD SCEQVALGLD NGQTRGLWHG WINFIFGDQP ALAQGAIEP EPRPALIAIG SAHLDPKLKA AFQGKKQLQE AIAFMKPGVA IDAITGTAKK DFLRFEEVVR LGAKLTAEVE L NLPDNLSE ...String: MRGIEITITM QSDWHVGTGM GRGELDSVVQ RDGDNLPYIP GKTLTGILRD SCEQVALGLD NGQTRGLWHG WINFIFGDQP ALAQGAIEP EPRPALIAIG SAHLDPKLKA AFQGKKQLQE AIAFMKPGVA IDAITGTAKK DFLRFEEVVR LGAKLTAEVE L NLPDNLSE TNKKVIAGIL ASGAKLTERL GGKRRRGNGR CELKFSGYSD QQIQWLKDNY QSVDQPPKYQ QNKLQSAGDN PE QQPPWHI IPLTIKTLSP VVLPARTVGN VVECLDYIPG RYLLGYIHKT LGEYFDVSQA IAAGDLIITN ATIKIDGKAG RAT PFCLFG EKLDGGLGKG KGVYNRFQES EPDGIQLKGE RGGYVGQFEQ EQRNLPNTGK INSELFTHNT IQDDVQRPTS DVGG VYSYE AIIAGQTFVA ELRLPDSLVK QITSKNKNWQ AQLKATIRIG QSKKDQYGKI EVTSGNSADL PKPTGNNKTL SIWFL SDIL LRGDRLNFNA TPDDLKKYLE NALDIKLKER SDNDLICIAL RSQRTESWQV RWGLPRPSLV GWQAGSCLIY DIESGT VNA EKLQELMITG IGDRCTEGYG QIGFNDPLLS ASLGKLTAKP KASNNQSQNS QSNPLPTNHP TQDYARLIEK AAWREAI QN KALALASSRA KREEILGIKI MGKDSQPTMT QLGGFRSVLK RLHSRNNRDI VTGYLTALEQ VSNRKEKWSN TSQGLTKI R NLVTQENLIW NHLDIDFSPL TITQNGVNQL KSELWAEAVR TLVDAIIRGH KRDLEKAQEN ESNQQSQGAA UniProtKB: CRISPR type III-associated protein domain-containing protein |
-Macromolecule #2: TIGR03984 family CRISPR-associated protein
| Macromolecule | Name: TIGR03984 family CRISPR-associated protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.899844 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPAGGRLMKN LYHYHQYEIT LESAVDSCKN HLQAAIGLLY SPQKCELVKL DNSGKLVDSY NRLKFNNLGV FEARFFNLNC ELRWVNESN GNGTAVLLSE SDITLTGFEK GLQEFITAID QQYLLWGEPA KHPPNADGWQ RLAEARIGKL DIPLDNPLKP K DRVFLTSE ...String: MPAGGRLMKN LYHYHQYEIT LESAVDSCKN HLQAAIGLLY SPQKCELVKL DNSGKLVDSY NRLKFNNLGV FEARFFNLNC ELRWVNESN GNGTAVLLSE SDITLTGFEK GLQEFITAID QQYLLWGEPA KHPPNADGWQ RLAEARIGKL DIPLDNPLKP K DRVFLTSE EYIAEVDDFG NCAVIDERLI KLEVK UniProtKB: TIGR03984 family CRISPR-associated protein |
-Macromolecule #3: Cas10
| Macromolecule | Name: Cas10 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 64.335309 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAHHHHHHVG TENLYFQGFL VLIETSGNQH FIFSTNKLRE NIGASELTYL ATTEILFQGV DRVFQTNYYD QWSDTNSLNF LADSKLNPA IDDPKNNADI EILLATSGKA IALVKEEGKA KQLIKEVTKQ ALINAPGLEI GGIYVNCNWQ DKLGVAKAVK E AHKQFEVN ...String: MAHHHHHHVG TENLYFQGFL VLIETSGNQH FIFSTNKLRE NIGASELTYL ATTEILFQGV DRVFQTNYYD QWSDTNSLNF LADSKLNPA IDDPKNNADI EILLATSGKA IALVKEEGKA KQLIKEVTKQ ALINAPGLEI GGIYVNCNWQ DKLGVAKAVK E AHKQFEVN RAKRAGANGR FLRLPIAAGC SVSELPASDF DYNADGDKIP VSTVSKVKRE TAKSAKKRLR SVDGRLVNDL AQ LEKSFDE LDWLAVVHAD GNGLGQILLS LEKYIGEQTN RNYIDKYRRL SLALDNCTIN AFKMAIAVFK EDSKKIDLPI VPL ILGGDD LTVICRGDYA LEFTREFLEA FEGQTETHDD IKVIAQKAFG VDRLSACAGI SIIKPHFPFS VAYTLAERLI KSAK EVKQK VTVTNSSPIT PFPCSAIDFH ILYDSSGIDF DRIREKLRPE DNTELYNRPY VVTAAENLSQ AQGYEWSQAH SLQTL ADRV SYLRSEDGEG KSALPSSQSH ALRTALYLEK NEADAQYSLI SQRYKILKNF AEDGENKSLF HLENGKYVTR FLDALD AKD FFANANHKNQ GE UniProtKB: Cas10/Cmr2 second palm domain-containing protein |
-Macromolecule #4: Cas7-2x
| Macromolecule | Name: Cas7-2x / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56.820832 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARKVTTRWK ITGTLIAETP LHIGGVGGDA DTDLALAVNG AGEYYVPGTS LAGALRGWMT QLLNNDESQI KDLWGDHLDA KRGASFVIV DDAVIHIPNN ADVEIREGVG IDRHFGTAAN GFKYSRAVIP KGSKFKLPLT FDSQDDGLPN ALIQLLCALE A GDIRLGAA ...String: MARKVTTRWK ITGTLIAETP LHIGGVGGDA DTDLALAVNG AGEYYVPGTS LAGALRGWMT QLLNNDESQI KDLWGDHLDA KRGASFVIV DDAVIHIPNN ADVEIREGVG IDRHFGTAAN GFKYSRAVIP KGSKFKLPLT FDSQDDGLPN ALIQLLCALE A GDIRLGAA KTRGLGRIKL DDLKLKSFAL DKPEGIFSAL LDQGKKLDWN QLKANVTYQS PPYLGISITW NPKDPVMVKA EG DGLAIDI LPLVSQVGSD VRFVIPGSSI KGILRTQAER IIRTICQSNG SEKNFLEQLR INLVNELFGS ASLSQKQNGK DID LGKIGA LAVNDCFSSL SMTPDQWKAV ENATEMTGNL QPALKQATGY PNNISQAYKV LQPAMHVAVD RWTGGAAEGM LYSV LEPIG VTWEPIQVHL DIARLKNYYH GKEEKLKPAI ALLLLVLRDL ANKKIPVGYG TNRGMGTITV SQITLNGKAL PTELE PLNK TMTCPNLTDL DEAFRQDLST AWKEWIADPI DLCQQEAA UniProtKB: CRISPR type III-associated protein domain-containing protein |
-Macromolecule #5: TIGR03986 family CRISPR-associated RAMP protein
| Macromolecule | Name: TIGR03986 family CRISPR-associated RAMP protein / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 90.334984 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTVGTLGVVG SAKNLKLQLS FINTRQQYVQ ITLFERNSFK VAEEEFSTEL VEIIKTALPT LKNKKVEFEE DGDQIKQIRE KGQAWVGAA EQIAPYVLPS GNITETPRNV NASNFHNPYN FVPALPRDGI TGDLGDCAPA GHSYYHGDKY SGRIAVKLTT V TPLLIPDA ...String: MTVGTLGVVG SAKNLKLQLS FINTRQQYVQ ITLFERNSFK VAEEEFSTEL VEIIKTALPT LKNKKVEFEE DGDQIKQIRE KGQAWVGAA EQIAPYVLPS GNITETPRNV NASNFHNPYN FVPALPRDGI TGDLGDCAPA GHSYYHGDKY SGRIAVKLTT V TPLLIPDA SKEEINNNHK TYPVRIGKDG KPYLPPTSIK GMLRSAYEAV TNSRLAVFED HDSRLAYRMP ATMGLQMVPA RI EGDNIVL YPGTSRIGNN GRPANNDPMY AAWLPYYQNR IAYDGSRDYQ MAEHGDHVRF WAERYTRGNF CYWRVRQIAR HNQ NLGNRP ERGRNYGQHH STGVIEQFEG FVYKTNKNIG NKHDERVFII DRESIEIPLS RDLRRKWREL ITSYQEIHKK EVDR GDTGP SAVNGAVWSR QIIADESERN LSDGTLCYAH VKKEDGQYKI LNLYPVMITR GLYEIAPVDL LDETLKPATD KKQLS PADR VFGWVNQRGN GCYKGQLRIH SVTCQHDDAI DDFGNQNFSV PLAILGQPKP EQARFYCADD RKGIPLEDGY DRDDGY SDS EQGLRGRKVY PHHKGLPNGY WSNPTEDRSQ QAIQGHYQEY RRPKKDGLEQ RDDQNRSVKG WVKPLTEFTF EIDVTNL SE VELGALLWLL TLPDLHFHRL GGGKPLGFGS VRLDIDPDKT DLRNGAGWRD YYGSLLETSQ PDFTTLISQW INAFQTAV K EEYGSSSFDQ VTFIKASGQS LQGFHDNASI HYPRSTPEPK PDGEAFKWFV ANEKGRRLAL PALEKSQSFP IKPS UniProtKB: CRISPR type III-associated protein domain-containing protein |
-Macromolecule #6: CRISPR RNA
| Macromolecule | Name: CRISPR RNA / type: rna / ID: 6 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.927167 KDa |
| Sequence | String: ACUGAAACUG UAGUAGAACC AAUCGGGGUC GUCAAUA |
-Macromolecule #7: Self-target RNA
| Macromolecule | Name: Self-target RNA / type: rna / ID: 7 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 19.265455 KDa |
| Sequence | String: CAUGACGGAU CGCGGGAGUU AUUGACGACC CCGAUUGGUU CUACUACAAA CGUGAUACUA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | Particles were monodisperse and homogeneous. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)