[English] 日本語
 Yorodumi
Yorodumi- EMDB-40229: Cryo-EM structure of octameric human CALHM1 with a I109W point mu... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of octameric human CALHM1 with a I109W point mutation | |||||||||
 Map data Map data | A map of the human CALHM1 protein (I109W point mutation), C8 symmetry | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | taste / assembly / calcium homeostasis modulator protein / membrane protein / channel / lipid binding / large-pore channel | |||||||||
| Function / homology |  Function and homology information Function and homology informationSensory perception of salty taste / sensory perception of bitter taste / ATP transport / sensory perception of umami taste / sensory perception of sweet taste / protein heterooligomerization / ATP export / voltage-gated monoatomic ion channel activity / regulation of monoatomic ion transmembrane transport / calcium-activated cation channel activity ...Sensory perception of salty taste / sensory perception of bitter taste / ATP transport / sensory perception of umami taste / sensory perception of sweet taste / protein heterooligomerization / ATP export / voltage-gated monoatomic ion channel activity / regulation of monoatomic ion transmembrane transport / calcium-activated cation channel activity / plasma membrane raft / monoatomic cation transport / monoatomic cation channel activity / voltage-gated calcium channel activity / protein homooligomerization / Sensory perception of sweet, bitter, and umami (glutamate) taste / basolateral plasma membrane / endoplasmic reticulum membrane / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
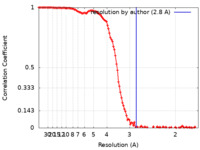
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Syrjanen JL / Furukawa H | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure of human CALHM1 reveals key locations for channel regulation and blockade by ruthenium red. Authors: Johanna L Syrjänen / Max Epstein / Ricardo Gómez / Hiro Furukawa /  Abstract: Calcium homeostasis modulator 1 (CALHM1) is a voltage-dependent channel involved in neuromodulation and gustatory signaling. Despite recent progress in the structural biology of CALHM1, insights into ...Calcium homeostasis modulator 1 (CALHM1) is a voltage-dependent channel involved in neuromodulation and gustatory signaling. Despite recent progress in the structural biology of CALHM1, insights into functional regulation, pore architecture, and channel blockade remain limited. Here we present the cryo-EM structure of human CALHM1, revealing an octameric assembly pattern similar to the non-mammalian CALHM1s and the lipid-binding pocket conserved across species. We demonstrate by MD simulations that this pocket preferentially binds a phospholipid over cholesterol to stabilize its structure and regulate the channel activities. Finally, we show that residues in the amino-terminal helix form the channel pore that ruthenium red binds and blocks. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40229.map.gz emd_40229.map.gz | 97.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40229-v30.xml emd-40229-v30.xml emd-40229.xml emd-40229.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40229_fsc.xml emd_40229_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_40229.png emd_40229.png | 91.5 KB | ||
| Filedesc metadata |  emd-40229.cif.gz emd-40229.cif.gz | 5.5 KB | ||
| Others |  emd_40229_half_map_1.map.gz emd_40229_half_map_1.map.gz emd_40229_half_map_2.map.gz emd_40229_half_map_2.map.gz | 115.7 MB 115.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40229 http://ftp.pdbj.org/pub/emdb/structures/EMD-40229 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40229 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40229 | HTTPS FTP |
-Related structure data
| Related structure data |  8gmpMC  8gmqC  8gmrC  8s8zC  8s90C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40229.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40229.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A map of the human CALHM1 protein (I109W point mutation), C8 symmetry | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.856 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: A half-map of the human CALHM1 protein (I109W point mutation)
| File | emd_40229_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A half-map of the human CALHM1 protein (I109W point mutation) | ||||||||||||
| Projections & Slices |
| ||||||||||||
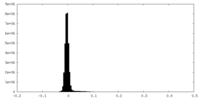
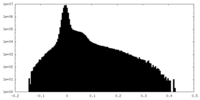
| Density Histograms |
-Half map: A half-map of the human CALHM1 protein (I109W point mutation)
| File | emd_40229_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A half-map of the human CALHM1 protein (I109W point mutation) | ||||||||||||
| Projections & Slices |
| ||||||||||||
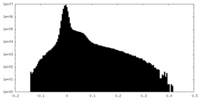
| Density Histograms |
- Sample components
Sample components
-Entire : Octameric human CALHM1 (I109W)
| Entire | Name: Octameric human CALHM1 (I109W) |
|---|---|
| Components |
|
-Supramolecule #1: Octameric human CALHM1 (I109W)
| Supramolecule | Name: Octameric human CALHM1 (I109W) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Calcium homeostasis modulator protein 1
| Macromolecule | Name: Calcium homeostasis modulator protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.842539 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MMDKFRMIFQ FLQSNQESFM NGICGIMALA SAQMYSAFDF NCPCLPGYNA AYSAGILLAP PLVLFLLGLV MNNNVSMLAE EWKRPPGRR AKDPAVLRYM FCSMAQRALW APVVWVAVTL LDGKCFLCAF CTAVPVSALG NGSLAPGLPA PELARLLARV P CPEIYDGD ...String: MMDKFRMIFQ FLQSNQESFM NGICGIMALA SAQMYSAFDF NCPCLPGYNA AYSAGILLAP PLVLFLLGLV MNNNVSMLAE EWKRPPGRR AKDPAVLRYM FCSMAQRALW APVVWVAVTL LDGKCFLCAF CTAVPVSALG NGSLAPGLPA PELARLLARV P CPEIYDGD WLLAREVAVR YLRCISQALG WSFVLLTTLL AFVVRSVRPC FTQAAFLKSK YWSHYIDIER KLFDETCTEH AK AFAKVCI QQFFEAMNHD LELGHTNGTL ATAPASAAAP TTPDGAEEER EKLRGITDQG TMNRLLGSAW SHPQFEK UniProtKB: Calcium homeostasis modulator protein 1 |
-Macromolecule #2: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 2 / Number of copies: 8 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)