[English] 日本語
 Yorodumi
Yorodumi- EMDB-40184: Structure of the Spizellomyces punctatus Fanzor (SpuFz) in comple... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Spizellomyces punctatus Fanzor (SpuFz) in complex with omega RNA and target DNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Fanzor / Eukaryotic / RNA-guided / nuclease / Gene editing / RNA BINDING PROTEIN / RNA BINDING PROTEIN-RNA-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationcarbohydrate transmembrane transporter activity / maltose binding / maltose transport / maltodextrin transmembrane transport / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / periplasmic space Similarity search - Function | |||||||||
| Biological species |  Spizellomyces punctatus (fungus) / synthetic construct (others) Spizellomyces punctatus (fungus) / synthetic construct (others) | |||||||||
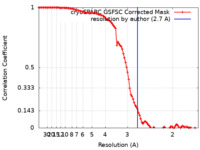
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Xu P / Saito M / Zhang F | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Fanzor is a eukaryotic programmable RNA-guided endonuclease. Authors: Makoto Saito / Peiyu Xu / Guilhem Faure / Samantha Maguire / Soumya Kannan / Han Altae-Tran / Sam Vo / AnAn Desimone / Rhiannon K Macrae / Feng Zhang /  Abstract: RNA-guided systems, which use complementarity between a guide RNA and target nucleic acid sequences for recognition of genetic elements, have a central role in biological processes in both ...RNA-guided systems, which use complementarity between a guide RNA and target nucleic acid sequences for recognition of genetic elements, have a central role in biological processes in both prokaryotes and eukaryotes. For example, the prokaryotic CRISPR-Cas systems provide adaptive immunity for bacteria and archaea against foreign genetic elements. Cas effectors such as Cas9 and Cas12 perform guide-RNA-dependent DNA cleavage. Although a few eukaryotic RNA-guided systems have been studied, including RNA interference and ribosomal RNA modification, it remains unclear whether eukaryotes have RNA-guided endonucleases. Recently, a new class of prokaryotic RNA-guided systems (termed OMEGA) was reported. The OMEGA effector TnpB is the putative ancestor of Cas12 and has RNA-guided endonuclease activity. TnpB may also be the ancestor of the eukaryotic transposon-encoded Fanzor (Fz) proteins, raising the possibility that eukaryotes are also equipped with CRISPR-Cas or OMEGA-like programmable RNA-guided endonucleases. Here we report the biochemical characterization of Fz, showing that it is an RNA-guided DNA endonuclease. We also show that Fz can be reprogrammed for human genome engineering applications. Finally, we resolve the structure of Spizellomyces punctatus Fz at 2.7 Å using cryogenic electron microscopy, showing the conservation of core regions among Fz, TnpB and Cas12, despite diverse cognate RNA structures. Our results show that Fz is a eukaryotic OMEGA system, demonstrating that RNA-guided endonucleases are present in all three domains of life. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40184.map.gz emd_40184.map.gz | 90.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40184-v30.xml emd-40184-v30.xml emd-40184.xml emd-40184.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40184_fsc.xml emd_40184_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_40184.png emd_40184.png | 85 KB | ||
| Others |  emd_40184_additional_1.map.gz emd_40184_additional_1.map.gz emd_40184_half_map_1.map.gz emd_40184_half_map_1.map.gz emd_40184_half_map_2.map.gz emd_40184_half_map_2.map.gz | 97.1 MB 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40184 http://ftp.pdbj.org/pub/emdb/structures/EMD-40184 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40184 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40184 | HTTPS FTP |
-Related structure data
| Related structure data |  8gkhMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40184.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40184.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_40184_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_40184_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_40184_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SpuFz-wRNA-tgDNA complex
| Entire | Name: SpuFz-wRNA-tgDNA complex |
|---|---|
| Components |
|
-Supramolecule #1: SpuFz-wRNA-tgDNA complex
| Supramolecule | Name: SpuFz-wRNA-tgDNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Spizellomyces punctatus (fungus) Spizellomyces punctatus (fungus) |
-Macromolecule #1: DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3')
| Macromolecule | Name: DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3') type: dna / ID: 1 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 4.594986 KDa |
| Sequence | String: (DC)(DG)(DG)(DT)(DA)(DC)(DC)(DC)(DG)(DG) (DG)(DC)(DA)(DT)(DA) |
-Macromolecule #3: DNA (35-MER)
| Macromolecule | Name: DNA (35-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 10.737919 KDa |
| Sequence | String: (DT)(DT)(DG)(DA)(DT)(DT)(DT)(DC)(DA)(DT) (DA)(DA)(DC)(DC)(DT)(DA)(DT)(DA)(DG)(DA) (DT)(DA)(DT)(DG)(DC)(DC)(DC)(DG)(DG) (DG)(DT)(DA)(DC)(DC)(DG) |
-Macromolecule #2: Maltodextrin-binding protein (Fragment),OrfB_Zn_ribbon domain-con...
| Macromolecule | Name: Maltodextrin-binding protein (Fragment),OrfB_Zn_ribbon domain-containing protein type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Spizellomyces punctatus (fungus) Spizellomyces punctatus (fungus) |
| Molecular weight | Theoretical: 118.029922 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKSSHHHHHH HHHHGSSMKI EEGKLVIWIN GDKGYNGLAE VGKKFEKDTG IKVTVEHPDK LEEKFPQVAA TGDGPDIIFW AHDRFGGYA QSGLLAEITP DKAFQDKLYP FTWDAVRYNG KLIAYPIAVE ALSLIYNKDL LPNPPKTWEE IPALDKELKA K GKSALMFN ...String: MKSSHHHHHH HHHHGSSMKI EEGKLVIWIN GDKGYNGLAE VGKKFEKDTG IKVTVEHPDK LEEKFPQVAA TGDGPDIIFW AHDRFGGYA QSGLLAEITP DKAFQDKLYP FTWDAVRYNG KLIAYPIAVE ALSLIYNKDL LPNPPKTWEE IPALDKELKA K GKSALMFN LQEPYFTWPL IAADGGYAFK YENGKYDIKD VGVDNAGAKA GLTFLVDLIK NKHMNADTDY SIAEAAFNKG ET AMTINGP WAWSNIDTSK VNYGVTVLPT FKGQPSKPFV GVLSAGINAA SPNKELAKEF LENYLLTDEG LEAVNKDKPL GAV ALKSYE EELAKDPRIA ATMENAQKGE IMPNIPQMSA FWYAVRTAVI NAASGRQTVD EALKDAQTGS ENLYFQSNAP PKKK QKLER LKKLDKPTLH TCNKTSFAKA FLPNETYRQR LLDYIAIIHQ LADHASHALK FYILSTSTSS FPVVHEDTIE AILYL LNKG EAWHPRKEAK KAWRDCLLPY VQRYCQIVGF IHPNLRGEQQ SINYLTVSMM TNLKVNVQEH FMQMLLRYIN LRFDVK GQK QRLPPKSDAR KAFFTRLRYL KSVFLFDVVP ELEFLDDLTP LESEVLEEIW SLDLPFLPND PLAYAIVADP MSFFPAY CK LSGLYEQYGF QRFSAIPLRR SLIQSHVRID TIILYQHILC ITRRDAETVE KDDLWMRVCN LCTKAFRSRC GMHFEGSI T TDGASVSVYL KHPEADKYGK RGARKSANTV AAEVKALYVE NNLPACRAAE NVVVIDPNKR DILYCQDSNG TTFRYTANQ RAVETGSRRF AKRREAMKEE AGVDLIESRI PSHKTMNLMD FTRYLLVRRA DWDRRKEFYS HPAHTRWKWH SFINRQKSES DLISNMRNK YGENFTVVMG DWSDAGRTAR FQTSSKTKGW RTLFKRNRID CFLLDEYKTS SVCPRCSSSE FVEKKFKTRP H SRPWRRRE GKIEKVHGLL GCTNPNCLQQ AWTSGMRYWN RDMLSTCNML LIVRSMLDGH GRPEVFSRSV PAVA UniProtKB: Maltodextrin-binding protein, Uncharacterized protein |
-Macromolecule #4: RNA (78-MER)
| Macromolecule | Name: RNA (78-MER) / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Spizellomyces punctatus (fungus) Spizellomyces punctatus (fungus) |
| Molecular weight | Theoretical: 25.050807 KDa |
| Sequence | String: UCCGAGCCGG UAGUCGCGCG GUUCAAUCCC UGGUGCGGGU GCUAGUGCAC CGGCUCCGCA CUAUCUAUAG GUUAUGAA |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 3 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 51.41 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)