+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Trypanosoma (MDH)4-(Pex5)2, close conformation | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Peroxisomal transport / Trypanosoma / Import / PEX / TRANSPORT PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationintracellular organelle lumen / peroxisome matrix targeting signal-1 binding / protein import into peroxisome matrix, docking / (S)-malate dehydrogenase (NAD+, oxaloacetate-forming) / L-malate dehydrogenase (NAD+) activity / carboxylic acid metabolic process / peroxisomal membrane / tricarboxylic acid cycle / cytosol / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.66 Å | ||||||||||||
 Authors Authors | Sonani RR / Artur B / Jemiola-Rzeminska M / Lipinski O / Patel SN / Sood T / Dubin G | ||||||||||||
| Funding support |  Poland, 3 items Poland, 3 items
| ||||||||||||
 Citation Citation |  Journal: Biorxiv / Year: 2023 Journal: Biorxiv / Year: 2023Title: Noncanonical interactions and conformational dynamics in cargo-Pex5-Pex14 ternary complex for peroxisomal import Authors: Sonani RR / Blat A / Jemiola-Rzeminska M / Lipinski O / Patel SN / Sood T / Dubin G | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40031.map.gz emd_40031.map.gz | 110.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40031-v30.xml emd-40031-v30.xml emd-40031.xml emd-40031.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40031.png emd_40031.png | 91.5 KB | ||
| Filedesc metadata |  emd-40031.cif.gz emd-40031.cif.gz | 6.4 KB | ||
| Others |  emd_40031_half_map_1.map.gz emd_40031_half_map_1.map.gz emd_40031_half_map_2.map.gz emd_40031_half_map_2.map.gz | 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40031 http://ftp.pdbj.org/pub/emdb/structures/EMD-40031 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40031 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40031 | HTTPS FTP |
-Related structure data
| Related structure data |  8gh2MC  8ggdC  8gghC  8gh3C  8gi0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40031.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40031.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
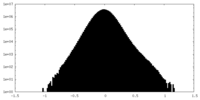
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_40031_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
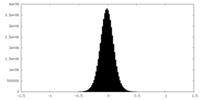
| Density Histograms |
-Half map: #1
| File | emd_40031_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
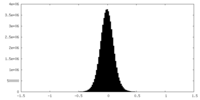
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of MDH-tetramer and PEX5
| Entire | Name: Complex of MDH-tetramer and PEX5 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of MDH-tetramer and PEX5
| Supramolecule | Name: Complex of MDH-tetramer and PEX5 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: malate dehydrogenase
| Macromolecule | Name: malate dehydrogenase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.106832 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVNVAVIGAA GGIGQSLSLL LLRELPFGST LSLYDVVGAP GVAADLSHID RAGITVKHAA GKLPPVPRDP ALTELAEGVD VFVIVAGVP RKPGMTRDDL FNVNAGIVMD LVLTCASVSP NACFCIVTNP VNSTTPIAAQ TLRKIGVYNK NKLLGVSLLD G LRATRFIN ...String: MVNVAVIGAA GGIGQSLSLL LLRELPFGST LSLYDVVGAP GVAADLSHID RAGITVKHAA GKLPPVPRDP ALTELAEGVD VFVIVAGVP RKPGMTRDDL FNVNAGIVMD LVLTCASVSP NACFCIVTNP VNSTTPIAAQ TLRKIGVYNK NKLLGVSLLD G LRATRFIN NARHPLVVPY VPVVGGHSDV TIVPLYSQIP GPLPDESTLK EIRKRVQVAG TEVVKAKAGR GSATLSMAEA GA RFTMHVV KALMGLDTPM VYAYVDTDGE HECPFLAMPV VLGKNGIERR LPIGPITTVE KEMLEEAVGV VKKNIAKGET FAR SKL UniProtKB: malate dehydrogenase |
-Macromolecule #2: Peroxisome targeting signal 1 receptor
| Macromolecule | Name: Peroxisome targeting signal 1 receptor / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 74.268422 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDCSTGAAIG QQFAKDAFHM HGGVGVGPTG NSEHDVLMNE MMMVQTPTGP AGEWTHQFAA YQGQQQQQQQ QHPQELAMRH QQNDAFMLR QQQEMEEAFC TFCTTHPHSH AHSHQPQGLV GPAMMGPQIM PPMMFGPGTG GFMMGAPPMM PYASMKFAGD A AMAAANNT ...String: MDCSTGAAIG QQFAKDAFHM HGGVGVGPTG NSEHDVLMNE MMMVQTPTGP AGEWTHQFAA YQGQQQQQQQ QHPQELAMRH QQNDAFMLR QQQEMEEAFC TFCTTHPHSH AHSHQPQGLV GPAMMGPQIM PPMMFGPGTG GFMMGAPPMM PYASMKFAGD A AMAAANNT NMTQGATATS TTSVQQELQQ QSSDNGWVEK LRDAEWAQDY SDAQVFTLEG QSEQTMEEHA KNSEFYQFMD KI RSKELLI DEETGQLVQG PGPDPDAPED AEYLKEWAAA EGLNMPPGFF EHMMQRPQGN NEQAEGRLFD GSNDALMDDG ALD NAADVE EWVREYAEAQ EQLQRVQNET NYPFEPNNPY MYHDKPMEEG IAMLQLANMA EAALAFEAVC QKEPENVEAW RRLG TTQAE NEKDCLAIIA LNHARMLDPK DIAVHAALAV SHTNEHNVGA ALQSLRSWLL SQPQYEHLGL VDLREVAADE GLDEV PEEN YFFAAPSEYR DCCTLLYAAV EMNPNDPQLH ASLGVLHNLS HRFDEAAKNF RRAVELRPDD AHMWNKLGAT LANGNR PQE ALEAYNRALD INPGYVRVMY NMAVSYSNMA QYPLAAKHIT RAIALQAGGT NPQGEGSRIA TRGLWDLLRM TLNLMDR SD LVEASWQQDL TPFLREFGLE EMAV UniProtKB: Peroxisome targeting signal 1 receptor |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 42.25 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.9 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)