[English] 日本語
 Yorodumi
Yorodumi- EMDB-3989: Morphology V - cross-beta amyloid fibril structure from the IGSNV... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3989 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Morphology V - cross-beta amyloid fibril structure from the IGSNVVTWYQQL peptide of AL immunoglobulin light chain by cryo-EM | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
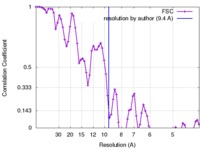
| Method | helical reconstruction / cryo EM / Resolution: 9.4 Å | |||||||||
 Authors Authors | Close W / Faendrich M | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: Physical basis of amyloid fibril polymorphism. Authors: William Close / Matthias Neumann / Andreas Schmidt / Manuel Hora / Karthikeyan Annamalai / Matthias Schmidt / Bernd Reif / Volker Schmidt / Nikolaus Grigorieff / Marcus Fändrich /   Abstract: Polymorphism is a key feature of amyloid fibril structures but it remains challenging to explain these variations for a particular sample. Here, we report electron cryomicroscopy-based ...Polymorphism is a key feature of amyloid fibril structures but it remains challenging to explain these variations for a particular sample. Here, we report electron cryomicroscopy-based reconstructions from different fibril morphologies formed by a peptide fragment from an amyloidogenic immunoglobulin light chain. The observed fibril morphologies vary in the number and cross-sectional arrangement of a structurally conserved building block. A comparison with the theoretically possible constellations reveals the experimentally observed spectrum of fibril morphologies to be governed by opposing sets of forces that primarily arise from the β-sheet twist, as well as peptide-peptide interactions within the fibril cross-section. Our results provide a framework for rationalizing and predicting the structure and polymorphism of cross-β fibrils, and suggest that a small number of physical parameters control the observed fibril architectures. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3989.map.gz emd_3989.map.gz | 48.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3989-v30.xml emd-3989-v30.xml emd-3989.xml emd-3989.xml | 10.2 KB 10.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3989_fsc.xml emd_3989_fsc.xml | 7.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_3989.png emd_3989.png | 51.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3989 http://ftp.pdbj.org/pub/emdb/structures/EMD-3989 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3989 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3989 | HTTPS FTP |
-Validation report
| Summary document |  emd_3989_validation.pdf.gz emd_3989_validation.pdf.gz | 220.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3989_full_validation.pdf.gz emd_3989_full_validation.pdf.gz | 219.7 KB | Display | |
| Data in XML |  emd_3989_validation.xml.gz emd_3989_validation.xml.gz | 8.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3989 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3989 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3989 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3989 | HTTPS FTP |
-Related structure data
| Related structure data |  3986C  3987C  3988C  3990C  3991C  3992C  3993C  3994C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3989.map.gz / Format: CCP4 / Size: 54.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3989.map.gz / Format: CCP4 / Size: 54.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Amyloidogenic Fragment IGSNVVTWYQQL of Immunoglobulin Light Chain...
| Entire | Name: Amyloidogenic Fragment IGSNVVTWYQQL of Immunoglobulin Light Chain of a Human AL Patient |
|---|---|
| Components |
|
-Supramolecule #1: Amyloidogenic Fragment IGSNVVTWYQQL of Immunoglobulin Light Chain...
| Supramolecule | Name: Amyloidogenic Fragment IGSNVVTWYQQL of Immunoglobulin Light Chain of a Human AL Patient type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism: synthetic construct (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 5.00 mg/mL |
|---|---|
| Buffer | pH: 8 / Component - Concentration: 50.0 mM / Component - Name: Tris |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 50 % / Instrument: GATAN CRYOPLUNGE 3 Details: Incubation of fibril solution on glow discharged holey carbon grid for 30 seconds and backside blotting for 4 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7420 pixel / Digitization - Dimensions - Height: 7676 pixel / Digitization - Frames/image: 1-73 / Number real images: 458 / Average exposure time: 21.9 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)