[English] 日本語
 Yorodumi
Yorodumi- EMDB-39753: Activation mechanism and novel binding site of the BKCa channel a... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Activation mechanism and novel binding site of the BKCa channel activator CTIBD | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | BKca channel / Slo1 / CTIBD / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | ||||||||||||
 Authors Authors | Lee N / Kim S / Jo H / Lee NY / Jin MS / Park CS | ||||||||||||
| Funding support |  Korea, Republic Of, 3 items Korea, Republic Of, 3 items
| ||||||||||||
 Citation Citation |  Journal: Life Sci Alliance / Year: 2024 Journal: Life Sci Alliance / Year: 2024Title: Activation mechanism and novel binding sites of the BK channel activator CTIBD. Authors: Narasaem Lee / Subin Kim / Na Young Lee / Heeji Jo / Pyeonghwa Jeong / Haushabhau S Pagire / Suvarna H Pagire / Jin Hee Ahn / Mi Sun Jin / Chul-Seung Park /   Abstract: The large-conductance calcium-activated potassium (BK) channel, which is crucial for urinary bladder smooth muscle relaxation, is a potential target for overactive bladder treatment. Our prior work ...The large-conductance calcium-activated potassium (BK) channel, which is crucial for urinary bladder smooth muscle relaxation, is a potential target for overactive bladder treatment. Our prior work unveiled CTIBD as a promising BK channel activator, altering and This study investigates CTIBD's activation mechanism, revealing its independence from the Ca and membrane voltage sensing of the BK channel. Cryo-electron microscopy disclosed that two CTIBD molecules bind to hydrophobic regions on the extracellular side of the lipid bilayer. Key residues (W22, W203, and F266) are important for CTIBD binding, and their replacement with alanine reduces CTIBD-mediated channel activation. The triple-mutant (W22A/W203A/F266A) channel showed the smallest shift with a minimal impact on activation and deactivation kinetics by CTIBD. At the single-channel level, CTIBD treatment was much less effective at increasing in the triple mutant, mainly because of a drastically increased dissociation rate compared with the WT. These findings highlight CTIBD's mechanism, offering crucial insights for developing small-molecule treatments for BK-related pathophysiological conditions. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39753.map.gz emd_39753.map.gz | 28.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39753-v30.xml emd-39753-v30.xml emd-39753.xml emd-39753.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_39753.png emd_39753.png | 108.8 KB | ||
| Filedesc metadata |  emd-39753.cif.gz emd-39753.cif.gz | 7.2 KB | ||
| Others |  emd_39753_half_map_1.map.gz emd_39753_half_map_1.map.gz emd_39753_half_map_2.map.gz emd_39753_half_map_2.map.gz | 27.9 MB 27.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39753 http://ftp.pdbj.org/pub/emdb/structures/EMD-39753 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39753 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39753 | HTTPS FTP |
-Related structure data
| Related structure data |  8z3sMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_39753.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39753.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.41 Å | ||||||||||||||||||||||||||||||||||||
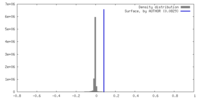
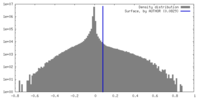
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_39753_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
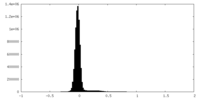
| Density Histograms |
-Half map: #1
| File | emd_39753_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
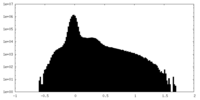
| Density Histograms |
- Sample components
Sample components
-Entire : Calcium-activated potassium channel subunit alpha-1 (Slo1)
| Entire | Name: Calcium-activated potassium channel subunit alpha-1 (Slo1) |
|---|---|
| Components |
|
-Supramolecule #1: Calcium-activated potassium channel subunit alpha-1 (Slo1)
| Supramolecule | Name: Calcium-activated potassium channel subunit alpha-1 (Slo1) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 500 KDa |
-Macromolecule #1: Calcium-activated potassium channel subunit alpha-1
| Macromolecule | Name: Calcium-activated potassium channel subunit alpha-1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 125.541461 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDALIIPVTM EVPCDSRGQR MWWAFLASSM VTFFGGLFII LLWRTLKYLW TVCCHCGGKT KEAQKINNGS SQADGTLKPV DEKEEAVAA EVGWMTSVKD WAGVMISAQT LTGRVLVVLV FALSIGALVI YFIDSSNPIE SCQNFYKDFT LQIDMAFNVF F LLYFGLRF ...String: MDALIIPVTM EVPCDSRGQR MWWAFLASSM VTFFGGLFII LLWRTLKYLW TVCCHCGGKT KEAQKINNGS SQADGTLKPV DEKEEAVAA EVGWMTSVKD WAGVMISAQT LTGRVLVVLV FALSIGALVI YFIDSSNPIE SCQNFYKDFT LQIDMAFNVF F LLYFGLRF IAANDKLWFW LEVNSVVDFF TVPPVFVSVY LNRSWLGLRF LRALRLIQFS EILQFLNILK TSNSIKLVNL LS IFISTWL TAAGFIHLVE NSGDPWENFQ NNQALTYWEC VYLLMVTMST VGYGDVYAKT TLGRLFMVFF ILGGLAMFAS YVP EIIELI GNRKKYGGSY SAVSGRKHIV VCGHITLESV SNFLKDFLHK DRDDVNVEIV FLHNISPNLE LEALFKRHFT QVEF YQGSV LNPHDLARVK IESADACLIL ANKYCADPDA EDASNIMRVI SIKNYHPKIR IITQMLQYHN KAHLLNIPSW NWKEG DDAI CLAELKLGFI AQSCLAQGLS TMLANLFSMR SFIKIEEDTW QKYYLEGVSN EMYTEYLSSA FVGLSFPTVC ELCFVK LKL LMIAIEYKSA NRESRILINP GNHLKIQEGT LGFFIASDAK EVKRAFFYCK ACHDDITDPK RIKKCGCKRP KMSIYKR MR RACCFDCGRS ERDCSCMSGR VRGNVDTLER AFPLSSVSVN DCSTSFRAFE DEQPSTLSPK KKQRNGGMRN SPNTSPKL M RHDPLLIPGN DQIDNMDSNV KKYDSTGMFH WCAPKEIEKV ILTRSEAAMT VLSGHVVVCI FGDVSSALIG LRNLVMPLR ASNFHYHELK HIVFVGSIEY LKREWETLHN FPKVSILPGT PLSRADLRAV NINLCDMCVI LSANQNNIDD TSLQDKECIL ASLNIKSMQ FDDSIGVLQA NSQGFTPPGM DRSSPDNSPV HGMLRQPSIT TGVNIPIITE LVNDTNVQFL DQDDDDDPDT E LYLTQPFA CGTAFAVSVL DSLMSATYFN DNILTLIRTL VTGGATPELE ALIAEENALR GGYSTPQTLA NRDRCRVAQL AL LDGPFAD LGDGGCYGDL FCKALKTYNM LCFGIYRLRD AHLSTPSQCT KRYVITNPPY EFELVPTDLI FCLMQFD UniProtKB: Peptide chain release factor subunit 1 |
-Macromolecule #2: 4-[4-(4-chlorophenyl)-3-(trifluoromethyl)-1,2-oxazol-5-yl]benzene...
| Macromolecule | Name: 4-[4-(4-chlorophenyl)-3-(trifluoromethyl)-1,2-oxazol-5-yl]benzene-1,3-diol type: ligand / ID: 2 / Number of copies: 8 / Formula: A1L04 |
|---|---|
| Molecular weight | Theoretical: 355.696 Da |
-Macromolecule #3: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 3 / Number of copies: 4 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 5 / Number of copies: 8 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.5 mg/mL | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 4806 / Average exposure time: 13.67 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.4000000000000001 µm / Nominal magnification: 73000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)