+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of HKU1B RBD with TMPRSS2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HKU1B / RBD / TMPRSS2 / VIRAL PROTEIN/HYDROLASE / VIRAL PROTEIN-HYDROLASE complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationtransmembrane protease serine 2 / protein autoprocessing / Attachment and Entry / serine-type peptidase activity / viral translation / Induction of Cell-Cell Fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell ...transmembrane protease serine 2 / protein autoprocessing / Attachment and Entry / serine-type peptidase activity / viral translation / Induction of Cell-Cell Fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / proteolysis / extracellular exosome / extracellular region / nucleoplasm / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Candidatus Accumulibacter adiacens (bacteria) Candidatus Accumulibacter adiacens (bacteria) | |||||||||
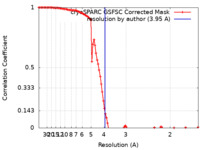
| Method | single particle reconstruction / cryo EM / Resolution: 3.95 Å | |||||||||
 Authors Authors | Gao X / Cui S / Ding W / Zhu K / Shang K / Zhu H | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2024 Journal: Cell Discov / Year: 2024Title: Structural basis for the interaction between human coronavirus HKU1 spike receptor binding domain and its receptor TMPRSS2. Authors: Xiaopan Gao / Kaixiang Zhu / Lin Wang / Kun Shang / Lei Hua / Bo Qin / Hongtao Zhu / Wei Ding / Sheng Cui /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39502.map.gz emd_39502.map.gz | 107.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39502-v30.xml emd-39502-v30.xml emd-39502.xml emd-39502.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_39502_fsc.xml emd_39502_fsc.xml | 10.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_39502.png emd_39502.png | 70.8 KB | ||
| Filedesc metadata |  emd-39502.cif.gz emd-39502.cif.gz | 6.5 KB | ||
| Others |  emd_39502_half_map_1.map.gz emd_39502_half_map_1.map.gz emd_39502_half_map_2.map.gz emd_39502_half_map_2.map.gz | 107.6 MB 107.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39502 http://ftp.pdbj.org/pub/emdb/structures/EMD-39502 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39502 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39502 | HTTPS FTP |
-Validation report
| Summary document |  emd_39502_validation.pdf.gz emd_39502_validation.pdf.gz | 732.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39502_full_validation.pdf.gz emd_39502_full_validation.pdf.gz | 731.8 KB | Display | |
| Data in XML |  emd_39502_validation.xml.gz emd_39502_validation.xml.gz | 18.3 KB | Display | |
| Data in CIF |  emd_39502_validation.cif.gz emd_39502_validation.cif.gz | 23.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39502 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39502 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39502 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39502 | HTTPS FTP |
-Related structure data
| Related structure data |  8yqqMC  8yoyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39502.map.gz / Format: CCP4 / Size: 115.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39502.map.gz / Format: CCP4 / Size: 115.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : HKU1B-RBD with TMPRSS2 receptor
| Entire | Name: HKU1B-RBD with TMPRSS2 receptor |
|---|---|
| Components |
|
-Supramolecule #1: HKU1B-RBD with TMPRSS2 receptor
| Supramolecule | Name: HKU1B-RBD with TMPRSS2 receptor / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Spike protein S1
| Macromolecule | Name: Spike protein S1 / type: protein_or_peptide / ID: 1 Details: Sequence reference for Candidatus Accumulibacter adiacens (2954378) is not available in UniProt at the time of biocuration. Current sequence reference is from UniProt ID Q0ZME7. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Candidatus Accumulibacter adiacens (bacteria) Candidatus Accumulibacter adiacens (bacteria) |
| Molecular weight | Theoretical: 31.703457 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: NLPDCDIDNW LNNVSVPSPL NWERRIFSNC NFNLSTLLRL VHVDSFSCNN LDKSKIFGSC FNSITVDKFA IPNRRRDDLQ LGSSGFLQS SNYKIDISSS SCQLYYSLPL VNVTINNFNP SSWNRRYGFG SFNLSSYDVV YSDHCFSVNS DFCPCADPSV V NSCAKSKP ...String: NLPDCDIDNW LNNVSVPSPL NWERRIFSNC NFNLSTLLRL VHVDSFSCNN LDKSKIFGSC FNSITVDKFA IPNRRRDDLQ LGSSGFLQS SNYKIDISSS SCQLYYSLPL VNVTINNFNP SSWNRRYGFG SFNLSSYDVV YSDHCFSVNS DFCPCADPSV V NSCAKSKP PSAICPAGTK YRHCDLDTTL YVKNWCRCSC LPDPISTYSP NTCPQKKVVV GIGEHCPGLG INEEKCGTQL NH SSCFCSP DAFLGWSFDS CISNNRCNIF SNFIFNGINS GTTCSNDL UniProtKB: Spike glycoprotein |
-Macromolecule #2: Transmembrane protease serine 2
| Macromolecule | Name: Transmembrane protease serine 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: transmembrane protease serine 2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 42.381848 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: MGSKCSNSGI ECDSSGTCIN PSNWCDGVSH CPGGEDENRC VRLYGPNFIL QVYSSQRKSW HPVCQDDWNE NYGRAACRDM GYKNNFYSS QGIVDDSGST SFMKLNTSAG NVDIYKKLYH SDACSSKAVV SLRCIACGVN LNSSRQSRIV GGESALPGAW P WQVSLHVQ ...String: MGSKCSNSGI ECDSSGTCIN PSNWCDGVSH CPGGEDENRC VRLYGPNFIL QVYSSQRKSW HPVCQDDWNE NYGRAACRDM GYKNNFYSS QGIVDDSGST SFMKLNTSAG NVDIYKKLYH SDACSSKAVV SLRCIACGVN LNSSRQSRIV GGESALPGAW P WQVSLHVQ NVHVCGGSII TPEWIVTAAH CVEKPLNNPW HWTAFAGILR QSFMFYGAGY QVEKVISHPN YDSKTKNNDI AL MKLQKPL TFNDLVKPVC LPNPGMMLQP EQLCWISGWG ATEEKGKTSE VLNAAKVLLI ETQRCNSRYV YDNLITPAMI CAG FLQGNV DSCQGDAGGP LVTSKNNIWW LIGDTSWGSG CAKAYRPGVY GNVMVFTDWI YRQMRADG UniProtKB: Transmembrane protease serine 2 |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)