[English] 日本語
 Yorodumi
Yorodumi- EMDB-39428: Structure of the Caspase-8/cFLIP death effector domain assembly -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Caspase-8/cFLIP death effector domain assembly | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | FADD / caspase-8 / cellular FLICE-like inhibitory protein / Death effector domain / APOPTOSIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of myoblast fusion / skeletal muscle atrophy / skeletal myofibril assembly / caspase-8 / syncytiotrophoblast cell differentiation involved in labyrinthine layer development / death effector domain binding / FasL/ CD95L signaling / TRAIL signaling / CD95 death-inducing signaling complex / regulation of skeletal muscle satellite cell proliferation ...negative regulation of myoblast fusion / skeletal muscle atrophy / skeletal myofibril assembly / caspase-8 / syncytiotrophoblast cell differentiation involved in labyrinthine layer development / death effector domain binding / FasL/ CD95L signaling / TRAIL signaling / CD95 death-inducing signaling complex / regulation of skeletal muscle satellite cell proliferation / Apoptotic execution phase / ripoptosome / Defective RIPK1-mediated regulated necrosis / Activation, myristolyation of BID and translocation to mitochondria / Microbial modulation of RIPK1-mediated regulated necrosis / TRAIL-activated apoptotic signaling pathway / TRIF-mediated programmed cell death / Regulation by c-FLIP / CASP8 activity is inhibited / Dimerization of procaspase-8 / regulation of necroptotic process / TLR3-mediated TICAM1-dependent programmed cell death / positive regulation of extracellular matrix organization / self proteolysis / Caspase activation via Death Receptors in the presence of ligand / positive regulation of macrophage differentiation / negative regulation of hepatocyte apoptotic process / positive regulation of glomerular mesangial cell proliferation / response to cobalt ion / positive regulation of hepatocyte proliferation / NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 / skeletal muscle tissue regeneration / death-inducing signaling complex / CLEC7A/inflammasome pathway / negative regulation of necroptotic process / regulation of tumor necrosis factor-mediated signaling pathway / tumor necrosis factor receptor binding / death receptor binding / natural killer cell activation / TNFR1-induced proapoptotic signaling / negative regulation of cellular response to transforming growth factor beta stimulus / negative regulation of cardiac muscle cell apoptotic process / RIPK1-mediated regulated necrosis / execution phase of apoptosis / response to anesthetic / Apoptotic cleavage of cellular proteins / regulation of innate immune response / response to testosterone / response to tumor necrosis factor / B cell activation / pyroptotic inflammatory response / macrophage differentiation / positive regulation of proteolysis / extrinsic apoptotic signaling pathway via death domain receptors / Caspase-mediated cleavage of cytoskeletal proteins / positive regulation of execution phase of apoptosis / cellular response to dexamethasone stimulus / negative regulation of extrinsic apoptotic signaling pathway via death domain receptors / skeletal muscle tissue development / negative regulation of reactive oxygen species biosynthetic process / extrinsic apoptotic signaling pathway / cellular response to nitric oxide / cysteine-type peptidase activity / regulation of cytokine production / : / cellular response to epidermal growth factor stimulus / T cell activation / negative regulation of canonical NF-kappaB signal transduction / positive regulation of interleukin-1 beta production / negative regulation of extrinsic apoptotic signaling pathway / protein maturation / Regulation of NF-kappa B signaling / cellular response to estradiol stimulus / apoptotic signaling pathway / Regulation of TNFR1 signaling / cellular response to mechanical stimulus / enzyme activator activity / positive regulation of neuron projection development / NOD1/2 Signaling Pathway / protein processing / Regulation of necroptotic cell death / cellular response to insulin stimulus / response to estradiol / peptidase activity / positive regulation of neuron apoptotic process / lamellipodium / heart development / cell body / protease binding / angiogenesis / scaffold protein binding / response to lipopolysaccharide / cellular response to hypoxia / response to ethanol / cytoskeleton / mitochondrial outer membrane / positive regulation of canonical NF-kappaB signal transduction / positive regulation of ERK1 and ERK2 cascade / positive regulation of cell migration / positive regulation of apoptotic process Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
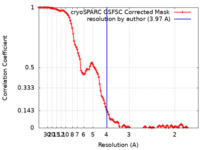
| Method | single particle reconstruction / cryo EM / Resolution: 3.97 Å | |||||||||
 Authors Authors | Lin S-C / Yang C-Y | |||||||||
| Funding support |  Taiwan, 1 items Taiwan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Reverse hierarchical DED assembly in the cFLIP-procaspase-8 and cFLIP-procaspase-8-FADD complexes. Authors: Chao-Yu Yang / Yi-Chun Tseng / Yi-Fan Tu / Bai-Jiun Kuo / Li-Chung Hsu / Chia-I Lien / You-Sheng Lin / Yin-Ting Wang / Yen-Chen Lu / Tsung-Wei Su / Yu-Chih Lo / Su-Chang Lin /  Abstract: cFLIP, a master anti-apoptotic regulator, targets the FADD-induced DED complexes of procaspase-8 in death receptor and ripoptosome signaling pathways. Several tumor cells maintain relatively high ...cFLIP, a master anti-apoptotic regulator, targets the FADD-induced DED complexes of procaspase-8 in death receptor and ripoptosome signaling pathways. Several tumor cells maintain relatively high levels of cFLIP in achieving their immortality. However, understanding the three-dimensional regulatory mechanism initiated or mediated by elevated levels of cFLIP has been limited by the absence of the atomic coordinates for cFLIP-induced DED complexes. Here we report the crystal plus cryo-EM structures to uncover an unconventional mechanism where cFLIP and procaspase-8 autonomously form a binary tandem DED complex, independent of FADD. This complex gains the ability to recruit FADD, thereby allosterically modulating cFLIP assembly and partially activating caspase-8 for RIPK1 cleavage. Our structure-guided mutagenesis experiments provide critical insights into these regulatory mechanisms, elucidating the resistance to apoptosis and necroptosis in achieving immortality. Finally, this research offers a unified model for the intricate bidirectional hierarchy-based processes using multiprotein helical assembly to govern cell fate decisions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39428.map.gz emd_39428.map.gz | 69.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39428-v30.xml emd-39428-v30.xml emd-39428.xml emd-39428.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_39428_fsc.xml emd_39428_fsc.xml | 10.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_39428.png emd_39428.png | 21 KB | ||
| Filedesc metadata |  emd-39428.cif.gz emd-39428.cif.gz | 6.2 KB | ||
| Others |  emd_39428_half_map_1.map.gz emd_39428_half_map_1.map.gz emd_39428_half_map_2.map.gz emd_39428_half_map_2.map.gz | 127.3 MB 127.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39428 http://ftp.pdbj.org/pub/emdb/structures/EMD-39428 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39428 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39428 | HTTPS FTP |
-Related structure data
| Related structure data |  8ynnMC  8ym4C  8ym5C  8ym6C  8yniC  8ynkC  8ynlC  8ynmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39428.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39428.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_39428_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_39428_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Caspase-8/cFLIP death effector domain assembly
| Entire | Name: Caspase-8/cFLIP death effector domain assembly |
|---|---|
| Components |
|
-Supramolecule #1: Caspase-8/cFLIP death effector domain assembly
| Supramolecule | Name: Caspase-8/cFLIP death effector domain assembly / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Caspase-8 subunit p10
| Macromolecule | Name: Caspase-8 subunit p10 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 55.191648 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDFSRNLYDI GEQLDSEDLA SLKFLSLDYI PQRKQEPIKD ALMLFQRLQE KRMLEESNLS FLKELLFRIN RLDLLITYLN TRKEEMERE LQTPGRAQIS AYRVMLYQIS EEVSRSELRS FKGGLQEEIS KCKLDDDMNL LDIFIEMEKR VILGEGKLDI L KRVCAQIN ...String: MDFSRNLYDI GEQLDSEDLA SLKFLSLDYI PQRKQEPIKD ALMLFQRLQE KRMLEESNLS FLKELLFRIN RLDLLITYLN TRKEEMERE LQTPGRAQIS AYRVMLYQIS EEVSRSELRS FKGGLQEEIS KCKLDDDMNL LDIFIEMEKR VILGEGKLDI L KRVCAQIN KSLLKIINDY EEFSKERSSS LEGSPDEFSN GEELCGVMTI SDSPREQDSE SQTLDKVYQM KSKPRGYCLI IN NHNFAKA REKVPKLHSI RDRNGTHLDA GALTTTFEEL HFEIKPHDDC TVEQIYEILK IYQLMDHSNM DCFICCILSH GDK GIIYGT DGQEAPIYEL TSQFTGLKCP SLAGKPKVFF IQAAQGDNYQ KGIPVETASE EQPYLEMALS SPQTRYIPDE ADFL LGMAT VNNCVSYRNP AEGTWYIQSL CQSLRERCPR GDDILTILTE VNYEVSNKDD KKNMGKQMPQ PTFTLRKKLV FPSD UniProtKB: Caspase-8 |
-Macromolecule #2: CASP8 and FADD-like apoptosis regulator subunit p43
| Macromolecule | Name: CASP8 and FADD-like apoptosis regulator subunit p43 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 20.878479 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSAEVIHQVE EALDTDEKEM LLFLCRDVAI DVVPPNVRDL LDILRERGKL SVGDLAELLY RVRRFDLLKR ILKMDRKAVE THLLRNPHL VSDYRVLMAE IGEDLDKSDV SSLIFLMKDY MGRGKISKEK SFLDLVVELE KLNLVAPDQL DLLEKCLKNI H RIDLKTKI QKYKQSVQGA GTS UniProtKB: CASP8 and FADD-like apoptosis regulator |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 279 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 72.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)