[English] 日本語
 Yorodumi
Yorodumi- EMDB-3942: Structure of the chloroplast ribosome with chl-RRF and hibernatio... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3942 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor (Focused refinement of LSU) | |||||||||
 Map data Map data | Postprocessed sharpened map that has been resampled onto the map used in the refinement of the coordinates | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Spinacia oleracea (spinach) Spinacia oleracea (spinach) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Perez Borema A / Aibara S / Paul B / Tobiasson V / Kimanius D / Forsberg BO / Wallden K / Lindahl E / Amunts A | |||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2018 Journal: Nat Plants / Year: 2018Title: Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor. Authors: Annemarie Perez Boerema / Shintaro Aibara / Bijoya Paul / Victor Tobiasson / Dari Kimanius / Björn O Forsberg / Karin Wallden / Erik Lindahl / A Amunts /   Abstract: Oxygenic photosynthesis produces oxygen and builds a variety of organic compounds, changing the chemistry of the air, the sea and fuelling the food chain on our planet. The photochemical reactions ...Oxygenic photosynthesis produces oxygen and builds a variety of organic compounds, changing the chemistry of the air, the sea and fuelling the food chain on our planet. The photochemical reactions underpinning this process in plants take place in the chloroplast. Chloroplasts evolved ~1.2 billion years ago from an engulfed primordial diazotrophic cyanobacterium, and chlororibosomes are responsible for synthesis of the core proteins driving photochemical reactions. Chlororibosomal activity is spatiotemporally coupled to the synthesis and incorporation of functionally essential co-factors, implying the presence of chloroplast-specific regulatory mechanisms and structural adaptation of the chlororibosome. Despite recent structural information, some of these aspects remained elusive. To provide new insights into the structural specialities and evolution, we report a comprehensive analysis of the 2.9-3.1 Å resolution electron cryo-microscopy structure of the spinach chlororibosome in complex with its recycling factor and hibernation-promoting factor. The model reveals a prominent channel extending from the exit tunnel to the chlororibosome exterior, structural re-arrangements that lead to increased surface area for translocon binding, and experimental evidence for parallel and convergent evolution of chloro- and mitoribosomes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3942.map.gz emd_3942.map.gz | 13.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3942-v30.xml emd-3942-v30.xml emd-3942.xml emd-3942.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3942.png emd_3942.png | 264.5 KB | ||
| Masks |  emd_3942_msk_1.map emd_3942_msk_1.map | 282.6 MB |  Mask map Mask map | |
| Others |  emd_3942_additional_1.map.gz emd_3942_additional_1.map.gz emd_3942_additional_2.map.gz emd_3942_additional_2.map.gz emd_3942_additional_3.map.gz emd_3942_additional_3.map.gz emd_3942_half_map_1.map.gz emd_3942_half_map_1.map.gz emd_3942_half_map_2.map.gz emd_3942_half_map_2.map.gz | 249.2 MB 25.9 MB 224.3 MB 225.1 MB 225.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3942 http://ftp.pdbj.org/pub/emdb/structures/EMD-3942 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3942 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3942 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3942.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3942.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed sharpened map that has been resampled onto the map used in the refinement of the coordinates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_3942_msk_1.map emd_3942_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
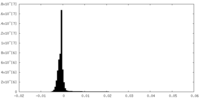
| Density Histograms |
-Additional map: unsharpened map that has been resampled onto the...
| File | emd_3942_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map that has been resampled onto the map used in the refinement of the coordinates | ||||||||||||
| Projections & Slices |
| ||||||||||||
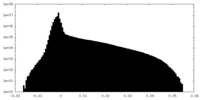
| Density Histograms |
-Additional map: Postprocessed sharpened map that has not been resampled...
| File | emd_3942_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessed sharpened map that has not been resampled onto the map used in the refinement of the coordinates | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: unsharpened map that has not been resampled onto...
| File | emd_3942_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map that has not been resampled onto the map used in the refinement of the coordinates | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: halfmap 1
| File | emd_3942_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
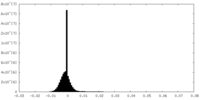
| Density Histograms |
-Half map: halfmap 2
| File | emd_3942_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Chloroplast ribosome in complex with RRF - Focussed refinement on LSU
| Entire | Name: Chloroplast ribosome in complex with RRF - Focussed refinement on LSU |
|---|---|
| Components |
|
-Supramolecule #1: Chloroplast ribosome in complex with RRF - Focussed refinement on LSU
| Supramolecule | Name: Chloroplast ribosome in complex with RRF - Focussed refinement on LSU type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Spinacia oleracea (spinach) Spinacia oleracea (spinach) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 4.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 130300 |
|---|---|
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)