+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | The Cryo-EM structure of IL-12, receptor subunit beta-1 and receptor subunit beta-2 complex, local refinement | |||||||||
 マップデータ マップデータ | A local refinement map of previous deposition of PDB ID 8XRP. | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | IL-12 / IL-12RB1 / IL-12RB2 / receptor complex / CYTOKINE | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報interleukin-27 binding / interleukin-12 beta subunit binding / late endosome lumen / interleukin-12 alpha subunit binding / interleukin-12 complex / interleukin-23 complex / natural killer cell activation involved in immune response / negative regulation of vascular endothelial growth factor signaling pathway / positive regulation of dendritic cell chemotaxis / positive regulation of natural killer cell activation ...interleukin-27 binding / interleukin-12 beta subunit binding / late endosome lumen / interleukin-12 alpha subunit binding / interleukin-12 complex / interleukin-23 complex / natural killer cell activation involved in immune response / negative regulation of vascular endothelial growth factor signaling pathway / positive regulation of dendritic cell chemotaxis / positive regulation of natural killer cell activation / positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target / negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis / positive regulation of lymphocyte proliferation / positive regulation of tissue remodeling / positive regulation of smooth muscle cell apoptotic process / positive regulation of NK T cell activation / positive regulation of T-helper 1 type immune response / sexual reproduction / positive regulation of mononuclear cell proliferation / positive regulation of natural killer cell mediated cytotoxicity / interleukin-12 receptor binding / interleukin-12 receptor complex / interleukin-23 receptor complex / T-helper cell differentiation / positive regulation of memory T cell differentiation / interleukin-23-mediated signaling pathway / Interleukin-23 signaling / positive regulation of T-helper 17 type immune response / interleukin-12-mediated signaling pathway / positive regulation of NK T cell proliferation / negative regulation of interleukin-17 production / positive regulation of osteoclast differentiation / Interleukin-12 signaling / Interleukin-35 Signalling / cytokine receptor activity / natural killer cell activation / positive regulation of granulocyte macrophage colony-stimulating factor production / response to UV-B / T-helper 1 type immune response / positive regulation of tyrosine phosphorylation of STAT protein / negative regulation of interleukin-10 production / defense response to protozoan / cytokine binding / Interleukin-10 signaling / positive regulation of interleukin-17 production / positive regulation of natural killer cell proliferation / positive regulation of activated T cell proliferation / positive regulation of interleukin-10 production / negative regulation of protein secretion / cell surface receptor signaling pathway via JAK-STAT / positive regulation of T-helper 17 cell lineage commitment / T cell proliferation / coreceptor activity / extrinsic apoptotic signaling pathway / positive regulation of defense response to virus by host / positive regulation of T cell proliferation / positive regulation of interleukin-12 production / positive regulation of cell adhesion / regulation of cytokine production / cytokine activity / growth factor activity / negative regulation of inflammatory response to antigenic stimulus / negative regulation of smooth muscle cell proliferation / positive regulation of non-canonical NF-kappaB signal transduction / cellular response to virus / cellular response to type II interferon / positive regulation of T cell mediated cytotoxicity / response to virus / positive regulation of type II interferon production / cytokine-mediated signaling pathway / positive regulation of inflammatory response / positive regulation of tumor necrosis factor production / cell migration / cellular response to lipopolysaccharide / Interleukin-4 and Interleukin-13 signaling / response to lipopolysaccharide / defense response to Gram-negative bacterium / defense response to virus / cell surface receptor signaling pathway / receptor complex / defense response to Gram-positive bacterium / immune response / endoplasmic reticulum lumen / protein heterodimerization activity / external side of plasma membrane / positive regulation of cell population proliferation / protein kinase binding / protein-containing complex binding / cell surface / signal transduction / extracellular space / extracellular region / identical protein binding / membrane / plasma membrane / cytosol 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.57 Å | |||||||||
 データ登録者 データ登録者 | Chen HQ / Ge XF | |||||||||
| 資金援助 |  中国, 1件 中国, 1件
| |||||||||
 引用 引用 |  ジャーナル: Structure / 年: 2024 ジャーナル: Structure / 年: 2024タイトル: Structure and assembly of the human IL-12 signaling complex. 著者: Huiqin Chen / Xiaofei Ge / Chun Li / Jianwei Zeng / Xinquan Wang /  要旨: Interleukin (IL)-12 is a heterodimeric pro-inflammatory cytokine. Our cryoelectron microscopy structure determination of human IL-12 in complex with IL-12Rβ1 and IL-12Rβ2 at a resolution of 3. ...Interleukin (IL)-12 is a heterodimeric pro-inflammatory cytokine. Our cryoelectron microscopy structure determination of human IL-12 in complex with IL-12Rβ1 and IL-12Rβ2 at a resolution of 3.75 Å reveals that IL-12Rβ2 primarily interacts with the IL-12p35 subunit via its N-terminal Ig-like domain, while IL-12Rβ1 binds to the p40 subunit with its N-terminal fibronectin III domain. This binding mode of IL-12 with its receptors is similar to that of IL-23 but shows notable differences with other cytokines. Through structural information and biochemical assays, we identified Y62, Y189, and K192 as key residues in IL-12p35, which bind to IL-12Rβ2 with high affinity and mediate IL-12 signal transduction. Furthermore, structural comparisons reveal two distinctive conformational states and structural plasticity of the heterodimeric interface in IL-12. As a result, our study advances our understanding of IL-12 signal initiation and opens up new opportunities for the engineering and therapeutic targeting of IL-12. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_39311.map.gz emd_39311.map.gz | 59.7 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-39311-v30.xml emd-39311-v30.xml emd-39311.xml emd-39311.xml | 18.7 KB 18.7 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_39311.png emd_39311.png | 73.3 KB | ||
| Filedesc metadata |  emd-39311.cif.gz emd-39311.cif.gz | 6.3 KB | ||
| その他 |  emd_39311_half_map_1.map.gz emd_39311_half_map_1.map.gz emd_39311_half_map_2.map.gz emd_39311_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39311 http://ftp.pdbj.org/pub/emdb/structures/EMD-39311 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39311 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39311 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_39311_validation.pdf.gz emd_39311_validation.pdf.gz | 875.3 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_39311_full_validation.pdf.gz emd_39311_full_validation.pdf.gz | 874.9 KB | 表示 | |
| XML形式データ |  emd_39311_validation.xml.gz emd_39311_validation.xml.gz | 12.3 KB | 表示 | |
| CIF形式データ |  emd_39311_validation.cif.gz emd_39311_validation.cif.gz | 14.4 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39311 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39311 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39311 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39311 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8yi7MC  8xrpC C: 同じ文献を引用 ( M: このマップから作成された原子モデル |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_39311.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_39311.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | A local refinement map of previous deposition of PDB ID 8XRP. | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.8697 Å | ||||||||||||||||||||||||||||||||||||
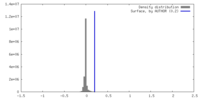
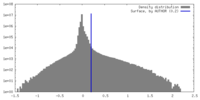
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: Half A map
| ファイル | emd_39311_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half A map | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
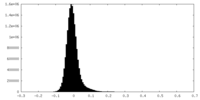
| 密度ヒストグラム |
-ハーフマップ: Half B map
| ファイル | emd_39311_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half B map | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
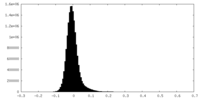
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : the ternary complex of IL-12 with IL-12R beta-1 and IL-12R beta-2
| 全体 | 名称: the ternary complex of IL-12 with IL-12R beta-1 and IL-12R beta-2 |
|---|---|
| 要素 |
|
-超分子 #1: the ternary complex of IL-12 with IL-12R beta-1 and IL-12R beta-2
| 超分子 | 名称: the ternary complex of IL-12 with IL-12R beta-1 and IL-12R beta-2 タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1-#4 |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 120 KDa |
-分子 #1: Interleukin-12 subunit alpha
| 分子 | 名称: Interleukin-12 subunit alpha / タイプ: protein_or_peptide / ID: 1 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 23.781508 KDa |
| 組換発現 | 生物種:  Baculovirus expression vector pFastBac1-HM (ウイルス) Baculovirus expression vector pFastBac1-HM (ウイルス) |
| 配列 | 文字列: LSLARNLPVA TPDPGMFPCL HHSQNLLRAV SNMLQKARQT LEFYPCTSEE IDHEDITKDK TSTVEACLPL ELTKNESCLN SRETSFITN GSCLASRKTS FMMALCLSSI YEDLKMYQVE FKTMNAKLLM DPKRQIFLDQ NMLAVIDELM QALNFNSETV P QKSSLEEP ...文字列: LSLARNLPVA TPDPGMFPCL HHSQNLLRAV SNMLQKARQT LEFYPCTSEE IDHEDITKDK TSTVEACLPL ELTKNESCLN SRETSFITN GSCLASRKTS FMMALCLSSI YEDLKMYQVE FKTMNAKLLM DPKRQIFLDQ NMLAVIDELM QALNFNSETV P QKSSLEEP DFYKTKIKLC ILLHAFRIRA VTIDRVMSYL NASHHHHHH UniProtKB: Interleukin-12 subunit alpha |
-分子 #2: Interleukin-12 subunit beta
| 分子 | 名称: Interleukin-12 subunit beta / タイプ: protein_or_peptide / ID: 2 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 34.811023 KDa |
| 組換発現 | 生物種:  Baculovirus expression vector pFastBac1-HM (ウイルス) Baculovirus expression vector pFastBac1-HM (ウイルス) |
| 配列 | 文字列: AIWELKKDVY VVELDWYPDA PGEMVVLTCD TPEEDGITWT LDQSSEVLGS GKTLTIQVKE FGDAGQYTCH KGGEVLSHSL LLLHKKEDG IWSTDILKDQ KEPKNKTFLR CEAKNYSGRF TCWWLTTIST DLTFSVKSSR GSSDPQGVTC GAATLSAERV R GDNKEYEY ...文字列: AIWELKKDVY VVELDWYPDA PGEMVVLTCD TPEEDGITWT LDQSSEVLGS GKTLTIQVKE FGDAGQYTCH KGGEVLSHSL LLLHKKEDG IWSTDILKDQ KEPKNKTFLR CEAKNYSGRF TCWWLTTIST DLTFSVKSSR GSSDPQGVTC GAATLSAERV R GDNKEYEY SVECQEDSAC PAAEESLPIE VMVDAVHKLK YENYTSSFFI RDIIKPDPPK NLQLKPLKNS RQVEVSWEYP DT WSTPHSY FSLTFCVQVQ GKSKREKKDR VFTDKTSATV ICRKNASISV RAQDRYYSSS WSEWASVPCS UniProtKB: Interleukin-12 subunit beta |
-分子 #3: Interleukin-12 receptor subunit beta-2
| 分子 | 名称: Interleukin-12 receptor subunit beta-2 / タイプ: protein_or_peptide / ID: 3 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 34.627477 KDa |
| 組換発現 | 生物種:  Baculovirus expression vector pFastBac1-HM (ウイルス) Baculovirus expression vector pFastBac1-HM (ウイルス) |
| 配列 | 文字列: KIDACKRGDV TVKPSHVILL GSTVNITCSL KPRQGCFHYS RRNKLILYKF DRRINFHHGH SLNSQVTGLP LGTTLFVCKL ACINSDEIQ ICGAEIFVGV APEQPQNLSC IQKGEQGTVA CTWERGRDTH LYTEYTLQLS GPKNLTWQKQ CKDIYCDYLD F GINLTPES ...文字列: KIDACKRGDV TVKPSHVILL GSTVNITCSL KPRQGCFHYS RRNKLILYKF DRRINFHHGH SLNSQVTGLP LGTTLFVCKL ACINSDEIQ ICGAEIFVGV APEQPQNLSC IQKGEQGTVA CTWERGRDTH LYTEYTLQLS GPKNLTWQKQ CKDIYCDYLD F GINLTPES PESNFTAKVT AVNSLGSSSS LPSTFTFLDI VRPLPPWDIR IKFQKASVSR CTLYWRDEGL VLLNRLRYRP SN SRLWNMV NVTKAKGRHD LLDLKPFTEY EFQISSKLHL YKGSWSDWSE SLRAQTPEEH HHHHH UniProtKB: Interleukin-12 receptor subunit beta-2 |
-分子 #4: Interleukin-12 receptor subunit beta-1
| 分子 | 名称: Interleukin-12 receptor subunit beta-1 / タイプ: protein_or_peptide / ID: 4 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 24.736449 KDa |
| 組換発現 | 生物種:  Baculovirus expression vector pFastBac1-HM (ウイルス) Baculovirus expression vector pFastBac1-HM (ウイルス) |
| 配列 | 文字列: CRTSECCFQD PPYPDADSGS ASGPRDLRCY RISSDRYECS WQYEGPTAGV SHFLRCCLSS GRCCYFAAGS ATRLQFSDQA GVSVLYTVT LWVESWARNQ TEKSPEVTLQ LYNSVKYEPP LGDIKVSKLA GQLRMEWETP DNQVGAEVQF RHRTPSSPWK L GDCGPQDD ...文字列: CRTSECCFQD PPYPDADSGS ASGPRDLRCY RISSDRYECS WQYEGPTAGV SHFLRCCLSS GRCCYFAAGS ATRLQFSDQA GVSVLYTVT LWVESWARNQ TEKSPEVTLQ LYNSVKYEPP LGDIKVSKLA GQLRMEWETP DNQVGAEVQF RHRTPSSPWK L GDCGPQDD DTESCLCPLE MNVAQEFQLR RRQLGSQGSS WSKWSSPVCV PPENPHHHHH H UniProtKB: Interleukin-12 receptor subunit beta-1 |
-分子 #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| 分子 | 名称: 2-acetamido-2-deoxy-beta-D-glucopyranose / タイプ: ligand / ID: 6 / コピー数: 1 / 式: NAG |
|---|---|
| 分子量 | 理論値: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 8 構成要素:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| グリッド | モデル: Quantifoil R1.2/1.3 / 材質: GOLD / メッシュ: 300 | |||||||||
| 凍結 | 凍結剤: ETHANE / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.0 µm / 最小 デフォーカス(公称値): 0.5 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
- 画像解析
画像解析
| 初期モデル | モデルのタイプ: NONE |
|---|---|
| 最終 再構成 | 解像度のタイプ: BY AUTHOR / 解像度: 3.57 Å / 解像度の算出法: FSC 0.143 CUT-OFF / 使用した粒子像数: 332015 |
| 初期 角度割当 | タイプ: ANGULAR RECONSTITUTION |
| 最終 角度割当 | タイプ: ANGULAR RECONSTITUTION |
 ムービー
ムービー コントローラー
コントローラー













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)