+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | FluPol-NS2-3 complex (hexamer) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PROTEIN BINDING | |||||||||
| Biological species |  Influenza A virus (A/Aichi/2/1968(H3N2)) Influenza A virus (A/Aichi/2/1968(H3N2)) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.74 Å | |||||||||
 Authors Authors | Peng Q / Sun JQ | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: EMBO Rep / Year: 2024 Journal: EMBO Rep / Year: 2024Title: NS2 induces an influenza A RNA polymerase hexamer and acts as a transcription to replication switch. Authors: Junqing Sun / Lu Kuai / Lei Zhang / Yufeng Xie / Yanfang Zhang / Yan Li / Qi Peng / Yuekun Shao / Qiuxian Yang / Wen-Xia Tian / Junhao Zhu / Jianxun Qi / Yi Shi / Tao Deng / George F Gao /  Abstract: Genome transcription and replication of influenza A virus (FluA), catalyzed by viral RNA polymerase (FluAPol), are delicately controlled across the virus life cycle. A switch from transcription to ...Genome transcription and replication of influenza A virus (FluA), catalyzed by viral RNA polymerase (FluAPol), are delicately controlled across the virus life cycle. A switch from transcription to replication occurring at later stage of an infection is critical for progeny virion production and viral non-structural protein NS2 has been implicated in regulating the switch. However, the underlying regulatory mechanisms and the structure of NS2 remained elusive for years. Here, we determine the cryo-EM structure of the FluAPol-NS2 complex at ~3.0 Å resolution. Surprisingly, three domain-swapped NS2 dimers arrange three symmetrical FluPol dimers into a highly ordered barrel-like hexamer. Further structural and functional analyses demonstrate that NS2 binding not only hampers the interaction between FluAPol and the Pol II CTD because of steric conflicts, but also impairs FluAPol transcriptase activity by stalling it in the replicase conformation. Moreover, this is the first visualization of the full-length NS2 structure. Our findings uncover key molecular mechanisms of the FluA transcription-replication switch and have implications for the development of antivirals. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39251.map.gz emd_39251.map.gz | 587.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39251-v30.xml emd-39251-v30.xml emd-39251.xml emd-39251.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_39251.png emd_39251.png | 33.7 KB | ||
| Filedesc metadata |  emd-39251.cif.gz emd-39251.cif.gz | 3.8 KB | ||
| Others |  emd_39251_half_map_1.map.gz emd_39251_half_map_1.map.gz emd_39251_half_map_2.map.gz emd_39251_half_map_2.map.gz | 622.1 MB 622.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39251 http://ftp.pdbj.org/pub/emdb/structures/EMD-39251 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39251 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39251 | HTTPS FTP |
-Validation report
| Summary document |  emd_39251_validation.pdf.gz emd_39251_validation.pdf.gz | 855.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39251_full_validation.pdf.gz emd_39251_full_validation.pdf.gz | 855.4 KB | Display | |
| Data in XML |  emd_39251_validation.xml.gz emd_39251_validation.xml.gz | 19.3 KB | Display | |
| Data in CIF |  emd_39251_validation.cif.gz emd_39251_validation.cif.gz | 22.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39251 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39251 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39251 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39251 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_39251.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39251.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.89 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_39251_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
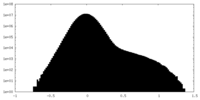
| Density Histograms |
-Half map: #2
| File | emd_39251_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
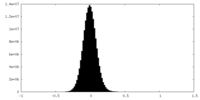
| Density Histograms |
- Sample components
Sample components
-Entire : FluPol-NS2 complex (hexamer)
| Entire | Name: FluPol-NS2 complex (hexamer) |
|---|---|
| Components |
|
-Supramolecule #1: FluPol-NS2 complex (hexamer)
| Supramolecule | Name: FluPol-NS2 complex (hexamer) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|
-Supramolecule #2: FluPol
| Supramolecule | Name: FluPol / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Aichi/2/1968(H3N2)) Influenza A virus (A/Aichi/2/1968(H3N2)) |
-Supramolecule #3: NS2
| Supramolecule | Name: NS2 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #4 |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Aichi/2/1968(H3N2)) Influenza A virus (A/Aichi/2/1968(H3N2)) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.001 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.74 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 351243 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)