[English] 日本語
 Yorodumi
Yorodumi- EMDB-39212: Cryo-EM structure of Dragon Grouper nervous necrosis virus-like p... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Dragon Grouper nervous necrosis virus-like particle at pH8.0 (3.23A) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nervous necrosis virus / Dragon Grouper / Cryo-EM structure / VIRUS | |||||||||
| Function / homology | Nodavirus capsid / nodavirus capsid protein / Viral coat protein subunit / viral capsid / Capsid protein alpha Function and homology information Function and homology information | |||||||||
| Biological species |  Dragon grouper nervous necrosis virus Dragon grouper nervous necrosis virus | |||||||||
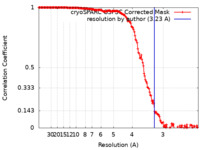
| Method | single particle reconstruction / cryo EM / Resolution: 3.23 Å | |||||||||
 Authors Authors | Wang CH / Chang WH | |||||||||
| Funding support |  Taiwan, 2 items Taiwan, 2 items
| |||||||||
 Citation Citation |  Journal: ACS Infect Dis / Year: 2024 Journal: ACS Infect Dis / Year: 2024Title: Molecular Mechanism of pH-Induced Protrusion Configuration Switching in Piscine Betanodavirus Implies a Novel Antiviral Strategy. Authors: Petra Štěrbová / Chun-Hsiung Wang / Kathleen J D Carillo / Yuan-Chao Lou / Takayuki Kato / Keiichi Namba / Der-Lii M Tzou / Wei-Hau Chang /   Abstract: Many viruses contain surface spikes or protrusions that are essential for virus entry. These surface structures can thereby be targeted by antiviral drugs to treat viral infections. Nervous necrosis ...Many viruses contain surface spikes or protrusions that are essential for virus entry. These surface structures can thereby be targeted by antiviral drugs to treat viral infections. Nervous necrosis virus (NNV), a simple nonenveloped virus in the genus of betanodavirus, infects fish and damages aquaculture worldwide. NNV has 60 conspicuous surface protrusions, each comprising three protrusion domains (P-domain) of its capsid protein. NNV uses protrusions to bind to common receptors of sialic acids on the host cell surface to initiate its entry via the endocytic pathway. However, structural alterations of NNV in response to acidic conditions encountered during this pathway remain unknown, while detailed interactions of protrusions with receptors are unclear. Here, we used cryo-EM to discover that Grouper NNV protrusions undergo low-pH-induced compaction and resting. NMR and molecular dynamics (MD) simulations were employed to probe the atomic details. A solution structure of the P-domain at pH 7.0 revealed a long flexible loop (amino acids 311-330) and a pocket outlined by this loop. Molecular docking analysis showed that the N-terminal moiety of sialic acid inserted into this pocket to interact with conserved residues inside. MD simulations demonstrated that part of this loop converted to a β-strand under acidic conditions, allowing for P-domain trimerization and compaction. Additionally, a low-pH-favored conformation is attained for the linker connecting the P-domain to the NNV shell, conferring resting protrusions. Our findings uncover novel pH-dependent conformational switching mechanisms underlying NNV protrusion dynamics potentially utilized for facilitating NNV entry, providing new structural insights into complex NNV-host interactions with the identification of putative druggable hotspots on the protrusion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39212.map.gz emd_39212.map.gz | 349.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39212-v30.xml emd-39212-v30.xml emd-39212.xml emd-39212.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_39212_fsc.xml emd_39212_fsc.xml | 15.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_39212.png emd_39212.png | 293.7 KB | ||
| Filedesc metadata |  emd-39212.cif.gz emd-39212.cif.gz | 5.3 KB | ||
| Others |  emd_39212_half_map_1.map.gz emd_39212_half_map_1.map.gz emd_39212_half_map_2.map.gz emd_39212_half_map_2.map.gz | 339.7 MB 339.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39212 http://ftp.pdbj.org/pub/emdb/structures/EMD-39212 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39212 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39212 | HTTPS FTP |
-Validation report
| Summary document |  emd_39212_validation.pdf.gz emd_39212_validation.pdf.gz | 976.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39212_full_validation.pdf.gz emd_39212_full_validation.pdf.gz | 975.9 KB | Display | |
| Data in XML |  emd_39212_validation.xml.gz emd_39212_validation.xml.gz | 24.4 KB | Display | |
| Data in CIF |  emd_39212_validation.cif.gz emd_39212_validation.cif.gz | 31.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39212 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39212 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39212 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39212 | HTTPS FTP |
-Related structure data
| Related structure data |  8yf6MC  8xidC  8yf7C  8yf8C  8yf9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_39212.map.gz / Format: CCP4 / Size: 371.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39212.map.gz / Format: CCP4 / Size: 371.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.16 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_39212_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_39212_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dragon Grouper nervous necrosis virus-like particle at pH8.0 (180...
| Entire | Name: Dragon Grouper nervous necrosis virus-like particle at pH8.0 (180 subunits) |
|---|---|
| Components |
|
-Supramolecule #1: Dragon Grouper nervous necrosis virus-like particle at pH8.0 (180...
| Supramolecule | Name: Dragon Grouper nervous necrosis virus-like particle at pH8.0 (180 subunits) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Dragon grouper nervous necrosis virus Dragon grouper nervous necrosis virus |
-Macromolecule #1: Capsid protein alpha
| Macromolecule | Name: Capsid protein alpha / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Dragon grouper nervous necrosis virus Dragon grouper nervous necrosis virus |
| Molecular weight | Theoretical: 37.128754 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVRKGEKKLA KPPTTKAANP QPRRRANNRR RSNRTDAPVS KASTVTGFGR GTNDVHLSGM SRISQAVLPA GTGTDGYVVV DATIVPDLL PRLGHAARIF QRYAVETLEF EIQPMCPANT GGGYVAGFLP DPTDNDHTFD ALQATRGAVV AKWWESRTVR P QYTRTLLW ...String: MVRKGEKKLA KPPTTKAANP QPRRRANNRR RSNRTDAPVS KASTVTGFGR GTNDVHLSGM SRISQAVLPA GTGTDGYVVV DATIVPDLL PRLGHAARIF QRYAVETLEF EIQPMCPANT GGGYVAGFLP DPTDNDHTFD ALQATRGAVV AKWWESRTVR P QYTRTLLW TSSGKEQRLT SPGRLILLCV GNNTDVVNVS VLCRWSVRLS VPSLETPEET TAPIMTQGSL YNDSLSTNDF KS ILLGSTP LDIAPDGAVF QLDRPLSIDY SLGTGDVDRA VYWHLKKFAG NAGTPAGWFR WGIWDNFNKT FTDGVAYYSD EQP RQILLP VGTVCTRVDS EN UniProtKB: Capsid protein alpha |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 3 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2100F |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.7600000000000002 µm / Nominal defocus min: 0.5700000000000001 µm |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)