[English] 日本語
 Yorodumi
Yorodumi- EMDB-38769: Structure of Platelet-activating factor receptor-G protein comple... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Platelet-activating factor receptor-G protein complex bound to platelet-activating factor | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / PTAFR / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationplatelet activating factor receptor activity / inositol trisphosphate biosynthetic process / lipopolysaccharide immune receptor activity / G protein-coupled purinergic nucleotide receptor activity / Class A/1 (Rhodopsin-like receptors) / Interleukin-10 signaling / tertiary granule membrane / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration ...platelet activating factor receptor activity / inositol trisphosphate biosynthetic process / lipopolysaccharide immune receptor activity / G protein-coupled purinergic nucleotide receptor activity / Class A/1 (Rhodopsin-like receptors) / Interleukin-10 signaling / tertiary granule membrane / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / response to prostaglandin E / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / secretory granule membrane / regulation of mitotic spindle organization / Regulation of insulin secretion / lipopolysaccharide binding / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / phospholipid binding / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / response to peptide hormone / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / chemotaxis / Interferon gamma signaling / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / GDP binding / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / G protein activity / GTPase binding / Ca2+ pathway / fibroblast proliferation / midbody / cell cortex / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / immune response / ciliary basal body / G protein-coupled receptor signaling pathway / inflammatory response / cell division / lysosomal membrane / GTPase activity / Neutrophil degranulation / synapse / centrosome / GTP binding / protein-containing complex binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Fan W / Xu Y / Zhuang Y / Xu HE | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2024 Journal: Cell Rep / Year: 2024Title: Molecular basis for the activation of PAF receptor by PAF. Authors: Wenjia Fan / Youwei Xu / Xinheng He / Ping Luo / Jingpeng Zhu / Junrui Li / Ruolan Wang / Qingning Yuan / Kai Wu / Wen Hu / Yuxi Zhao / Shiqi Xu / Xi Cheng / Yue Wang / H Eric Xu / Youwen Zhuang /  Abstract: Platelet-activating factor (PAF) is a potent phospholipid mediator crucial in multiple inflammatory and immune responses through binding and activating the PAF receptor (PAFR). However, drug ...Platelet-activating factor (PAF) is a potent phospholipid mediator crucial in multiple inflammatory and immune responses through binding and activating the PAF receptor (PAFR). However, drug development targeting the PAFR has been limited, partly due to an incomplete understanding of its activation mechanism. Here, we present a 2.9-Å structure of the PAF-bound PAFR-G complex. Structural and mutagenesis analyses unveil a specific binding mode of PAF, with the choline head forming cation-π interactions within PAFR hydrophobic pocket, while the alkyl tail penetrates deeply into an aromatic cleft between TM4 and TM5. Binding of PAF modulates conformational changes in key motifs of PAFR, triggering the outward movement of TM6, TM7, and helix 8 for G protein coupling. Molecular dynamics simulation suggests a membrane-side pathway for PAF entry into PAFR via the TM4-TM5 cavity. By providing molecular insights into PAFR signaling, this work contributes a foundation for developing therapeutic interventions targeting PAF signal axis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38769.map.gz emd_38769.map.gz | 79.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38769-v30.xml emd-38769-v30.xml emd-38769.xml emd-38769.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38769.png emd_38769.png | 38.2 KB | ||
| Filedesc metadata |  emd-38769.cif.gz emd-38769.cif.gz | 6.5 KB | ||
| Others |  emd_38769_additional_1.map.gz emd_38769_additional_1.map.gz emd_38769_half_map_1.map.gz emd_38769_half_map_1.map.gz emd_38769_half_map_2.map.gz emd_38769_half_map_2.map.gz | 6.9 MB 80.3 MB 79.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38769 http://ftp.pdbj.org/pub/emdb/structures/EMD-38769 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38769 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38769 | HTTPS FTP |
-Related structure data
| Related structure data |  8xydMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38769.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38769.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.824 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_38769_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_38769_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_38769_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : PTAFR-PAF G protein complex
| Entire | Name: PTAFR-PAF G protein complex |
|---|---|
| Components |
|
-Supramolecule #1: PTAFR-PAF G protein complex
| Supramolecule | Name: PTAFR-PAF G protein complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Platelet-activating factor receptor
| Macromolecule | Name: Platelet-activating factor receptor / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.24227 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MEPHDSSHMD SEFRYTLFPI VYSIIFVLGV IANGYVLWVF ARLYPCKKFN EIKIFMVNLT MADMLFLITL PLWIVYYQNQ GNWILPKFL CNVAGCLFFI NTYCSVAFLG VITYNRFQAV TRPIKTAQAN TRKRGISLSL VIWVAIVGAA SYFLILDSTN T VPDSAGSG ...String: MEPHDSSHMD SEFRYTLFPI VYSIIFVLGV IANGYVLWVF ARLYPCKKFN EIKIFMVNLT MADMLFLITL PLWIVYYQNQ GNWILPKFL CNVAGCLFFI NTYCSVAFLG VITYNRFQAV TRPIKTAQAN TRKRGISLSL VIWVAIVGAA SYFLILDSTN T VPDSAGSG NVTRCFEHYE KGSVPVLIIH IFIVFSFFLV FLIILFCNLV IIRTLLMQPV QQQRNAEVKR RALWMVCTVL AV FIICFVP HHVVQLPWTL AELGFQDSKF HQAINDAHQV TLCLLSTNCV LDPVIYCFLT KKFRKHLTEK FYSMRSSRKC SRA TTDTVT EVVVPFNQIP GNSLKN UniProtKB: Platelet-activating factor receptor |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.784301 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GSLLQSELDQ LRQEAEQLKN QIRDARKACA DATLSQITNN IDPVGRIQMR TRRTLRGHLA KIYAMHWGTD SRLLVSASQD GKLIIWDSY TTNKVHAIPL RSSWVMTCAY APSGNYVACG GLDNICSIYN LKTREGNVRV SRELAGHTGY LSCCRFLDDN Q IVTSSGDT ...String: GSLLQSELDQ LRQEAEQLKN QIRDARKACA DATLSQITNN IDPVGRIQMR TRRTLRGHLA KIYAMHWGTD SRLLVSASQD GKLIIWDSY TTNKVHAIPL RSSWVMTCAY APSGNYVACG GLDNICSIYN LKTREGNVRV SRELAGHTGY LSCCRFLDDN Q IVTSSGDT TCALWDIETG QQTTTFTGHT GDVMSLSLAP DTRLFVSGAC DASAKLWDVR EGMCRQTFTG HESDINAICF FP NGNAFAT GSDDATCRLF DLRADQELMT YSHDNIICGI TSVSFSKSGR LLLAGYDDFN CNVWDALKAD RAGVLAGHDN RVS CLGVTD DGMAVATGSW DSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.56375 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFC UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.313863 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GCTLSAEDKA AVERSKMIDR NLREDGEKAA REVKLLLLGA GESGKSTIVK QMKIIHEAGY SEEECKQYKA VVYSNTIQSI IAIIRAMGR LKIDFGDSAR ADDARQLFVL AGAAEEGFMT AELAGVIKRL WKDSGVQACF NRSREYQLND SAAYYLNDLD R IAQPNYIP ...String: GCTLSAEDKA AVERSKMIDR NLREDGEKAA REVKLLLLGA GESGKSTIVK QMKIIHEAGY SEEECKQYKA VVYSNTIQSI IAIIRAMGR LKIDFGDSAR ADDARQLFVL AGAAEEGFMT AELAGVIKRL WKDSGVQACF NRSREYQLND SAAYYLNDLD R IAQPNYIP TQQDVLRTRV KTTGIVETHF TFKDLHFKMF DVGAQRSERK KWIHCFEGVT AIIFCVALSD YDLVLAEDEE MN RMHESMK LFDSICNNKW FTDTSIILFL NKKDLFEEKI KKSPLTICYP EYAGSNTYEE AAAYIQCQFE DLNKRKDTKE IYT HFTCST DTKNVQFVFD AVTDVIIKNN LKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #5: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.277299 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL |
-Macromolecule #6: (2R)-2-(acetyloxy)-3-(hexadecyloxy)propyl 2-(trimethylammonio)eth...
| Macromolecule | Name: (2R)-2-(acetyloxy)-3-(hexadecyloxy)propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 6 / Number of copies: 1 / Formula: PFS |
|---|---|
| Molecular weight | Theoretical: 523.683 Da |
| Chemical component information | 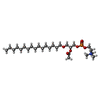 ChemComp-PFS: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: AlphaFold2 model |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 346106 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller
































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)












































