+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
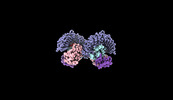
| Title | Cryo-EM structure of tomato NRC2 dimer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | tomato / helper NLR / dimer / PLANT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationdefense response to other organism / ADP binding / defense response to virus / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.84 Å | |||||||||
 Authors Authors | Sun Y / Ma SC / Chai JJ | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Oligomerization-mediated autoinhibition and cofactor binding of a plant NLR. Authors: Shoucai Ma / Chunpeng An / Aaron W Lawson / Yu Cao / Yue Sun / Eddie Yong Jun Tan / Jinheng Pan / Jan Jirschitzka / Florian Kümmel / Nitika Mukhi / Zhifu Han / Shan Feng / Bin Wu / Paul ...Authors: Shoucai Ma / Chunpeng An / Aaron W Lawson / Yu Cao / Yue Sun / Eddie Yong Jun Tan / Jinheng Pan / Jan Jirschitzka / Florian Kümmel / Nitika Mukhi / Zhifu Han / Shan Feng / Bin Wu / Paul Schulze-Lefert / Jijie Chai /    Abstract: Nucleotide-binding leucine-rich repeat (NLR) proteins play a pivotal role in plant immunity by recognizing pathogen effectors. Maintaining a balanced immune response is crucial, as excessive NLR ...Nucleotide-binding leucine-rich repeat (NLR) proteins play a pivotal role in plant immunity by recognizing pathogen effectors. Maintaining a balanced immune response is crucial, as excessive NLR expression can lead to unintended autoimmunity. Unlike most NLRs, the plant NLR required for cell death 2 (NRC2) belongs to a small NLR group characterized by constitutively high expression without self-activation. The mechanisms underlying NRC2 autoinhibition and activation are not yet understood. Here we show that Solanum lycopersicum (tomato) NRC2 (SlNRC2) forms dimers and tetramers and higher-order oligomers at elevated concentrations. Cryo-electron microscopy shows an inactive conformation of SlNRC2 in these oligomers. Dimerization and oligomerization not only stabilize the inactive state but also sequester SlNRC2 from assembling into an active form. Mutations at the dimeric or interdimeric interfaces enhance pathogen-induced cell death and immunity in Nicotiana benthamiana. The cryo-electron microscopy structures unexpectedly show inositol hexakisphosphate (IP) or pentakisphosphate (IP) bound to the inner surface of the C-terminal leucine-rich repeat domain of SlNRC2, as confirmed by mass spectrometry. Mutations at the inositol phosphate-binding site impair inositol phosphate binding of SlNRC2 and pathogen-induced SlNRC2-mediated cell death in N. benthamiana. Our study indicates a negative regulatory mechanism of NLR activation and suggests inositol phosphates as cofactors of NRCs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38679.map.gz emd_38679.map.gz | 64.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38679-v30.xml emd-38679-v30.xml emd-38679.xml emd-38679.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38679.png emd_38679.png | 23.8 KB | ||
| Filedesc metadata |  emd-38679.cif.gz emd-38679.cif.gz | 6.1 KB | ||
| Others |  emd_38679_half_map_1.map.gz emd_38679_half_map_1.map.gz emd_38679_half_map_2.map.gz emd_38679_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38679 http://ftp.pdbj.org/pub/emdb/structures/EMD-38679 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38679 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38679 | HTTPS FTP |
-Related structure data
| Related structure data |  8xuoMC  8xuqC  8xuvC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38679.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38679.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
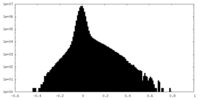
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_38679_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
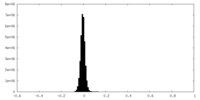
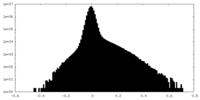
| Density Histograms |
-Half map: #1
| File | emd_38679_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
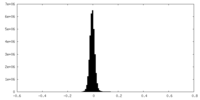
| Density Histograms |
- Sample components
Sample components
-Entire : NRC2 dimer
| Entire | Name: NRC2 dimer |
|---|---|
| Components |
|
-Supramolecule #1: NRC2 dimer
| Supramolecule | Name: NRC2 dimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: NRC2
| Macromolecule | Name: NRC2 / type: protein_or_peptide / ID: 1 Details: Sequence reference for NRC2 is not available in UniProt at the time of biocuration. Current sequence reference is from UniProt id A0A3Q7IF17. Regarding SlNRC2 with the accession number ...Details: Sequence reference for NRC2 is not available in UniProt at the time of biocuration. Current sequence reference is from UniProt id A0A3Q7IF17. Regarding SlNRC2 with the accession number Solyc10g047320 in the SGN database, here is the link: https://solgenomics.net/locus/36701/view. The source article for SlNRC2 is "Helper NLR proteins NRC2a/b and NRC3 but not NRC1 are required for Pto-mediated cell death and resistance in Nicotiana benthamiana" by Wu, Chih-Hang et al. The article was published in The New Phytologist, volume 209, issue 4, in 2016. The DOI is 10.1111/nph.13764. Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 86.267594 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DDDLSARGSE ERKPPVVEED DVVGFDEEAD IVINRLLGES NHLEVVPVVG MPGLGKTTLA NKIYKHPKIG YEFFTRIWVY VSQSYRRRE LFLNIISKFT RNTKQYHGMC EEDLADEIQE FLGKGGKYLV VLDDVWSDEA WERIKIAFPN NNKPNRVLLT T RDSKVAKQ ...String: DDDLSARGSE ERKPPVVEED DVVGFDEEAD IVINRLLGES NHLEVVPVVG MPGLGKTTLA NKIYKHPKIG YEFFTRIWVY VSQSYRRRE LFLNIISKFT RNTKQYHGMC EEDLADEIQE FLGKGGKYLV VLDDVWSDEA WERIKIAFPN NNKPNRVLLT T RDSKVAKQ CNPIPHDLKF LTEDESWILL EKKVFHKDKC PPELVLSGKS IAKKCKGLPL AIVVIAGALI GKGKTPREWK QV DDSVSEH LINRDHPENC NKLVQMSYDR LPYDLKACFL YCSAFPGGFQ IPAWKLIRLW IAEGFIQYKG HLSLECKGED NLN DLINRN LVMVMERTSD GQIKTCRLHD MLHEFCRQEA MKEENLFQEI KLGSEQYFPG KRELSTYRRL CIHSSVLDFF STKP SAEHV RSFLSFSSKK IEMPSADIPT IPKGFPLLRV LDVESINFSR FSREFYQLYH LRYVAFSSDS IKILPKLMGE LWNIQ TIII NTQQRTLDIQ ANIWNMERLR HLHTNSSAKL PVPVAPKNSK VTLVNQSLQT LSTIAPESCT EEVFARTPNL KKLGIR GKI SVLLDNKSAA SLKNVKRLEY LENLKLINDS SIQTSKLRLP PAYIFPTKLR KLTLLDTWLE WKDMSILGQL EHLEVLK MK ENGFSGESWE STGGFCSLLV LWIERTNLVS WKASADDFPR LKHLVLICCD NLKEVPIALA DIRSFQVMML QNSTKTAA I SARQIQAKKD NQTQQGTKNI AFKLSIFPPD L UniProtKB: NRC1 |
-Macromolecule #2: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #3: INOSITOL HEXAKISPHOSPHATE
| Macromolecule | Name: INOSITOL HEXAKISPHOSPHATE / type: ligand / ID: 3 / Number of copies: 2 / Formula: IHP |
|---|---|
| Molecular weight | Theoretical: 660.035 Da |
| Chemical component information |  ChemComp-IHP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.84 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 560935 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)