[English] 日本語
 Yorodumi
Yorodumi- EMDB-3864: Negative stain electron microscopy reconstruction of a cellulose ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3864 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
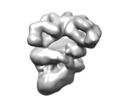
| Title | Negative stain electron microscopy reconstruction of a cellulose secretion (Bcs) macrocomplex from E. coli | |||||||||||||||
 Map data Map data | Bcs macrocomplex encompassing most inner-membrane and cytosolic components of the E. coli cellulose secretion system. Negative-stain EM | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  | |||||||||||||||
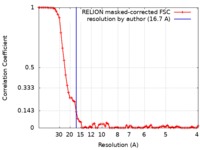
| Method | single particle reconstruction / negative staining / Resolution: 16.7 Å | |||||||||||||||
 Authors Authors | Krasteva PV / Fronzes R | |||||||||||||||
| Funding support |  France, 4 items France, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2017 Journal: Nat Commun / Year: 2017Title: Insights into the structure and assembly of a bacterial cellulose secretion system. Authors: Petya Violinova Krasteva / Joaquin Bernal-Bayard / Laetitia Travier / Fernando Ariel Martin / Pierre-Alexandre Kaminski / Gouzel Karimova / Rémi Fronzes / Jean-Marc Ghigo /   Abstract: Secreted exopolysaccharides present important determinants for bacterial biofilm formation, survival, and virulence. Cellulose secretion typically requires the concerted action of a c-di-GMP- ...Secreted exopolysaccharides present important determinants for bacterial biofilm formation, survival, and virulence. Cellulose secretion typically requires the concerted action of a c-di-GMP-responsive inner membrane synthase (BcsA), an accessory membrane-anchored protein (BcsB), and several additional Bcs components. Although the BcsAB catalytic duo has been studied in great detail, its interplay with co-expressed subunits remains enigmatic. Here we show that E. coli Bcs proteins partake in a complex protein interaction network. Electron microscopy reveals a stable, megadalton-sized macromolecular assembly, which encompasses most of the inner membrane and cytosolic Bcs components and features a previously unobserved asymmetric architecture. Heterologous reconstitution and mutational analyses point toward a structure-function model, where accessory proteins regulate secretion by affecting both the assembly and stability of the system. Altogether, these results lay the foundation for more comprehensive models of synthase-dependent exopolysaccharide secretion in biofilms and add a sophisticated secretory nanomachine to the diverse bacterial arsenal for virulence and adaptation. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3864.map.gz emd_3864.map.gz | 5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3864-v30.xml emd-3864-v30.xml emd-3864.xml emd-3864.xml | 13.6 KB 13.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3864_fsc.xml emd_3864_fsc.xml | 7.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_3864.png emd_3864.png | 48.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3864 http://ftp.pdbj.org/pub/emdb/structures/EMD-3864 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3864 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3864 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3864.map.gz / Format: CCP4 / Size: 35.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3864.map.gz / Format: CCP4 / Size: 35.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Bcs macrocomplex encompassing most inner-membrane and cytosolic components of the E. coli cellulose secretion system. Negative-stain EM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.9 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Protein macrocomplex encompassing most cytosolic and inner-membra...
| Entire | Name: Protein macrocomplex encompassing most cytosolic and inner-membrane components of the E. coli system for cellulose secretion |
|---|---|
| Components |
|
-Supramolecule #1: Protein macrocomplex encompassing most cytosolic and inner-membra...
| Supramolecule | Name: Protein macrocomplex encompassing most cytosolic and inner-membrane components of the E. coli system for cellulose secretion type: complex / ID: 1 / Parent: 0 Details: Protein macrocomplex encompassing cytosolic (BcsRQEF) and inner-membrane (BcsAB) components. Isolated from purified and detergent-solubilised E. coli 1094 membranes using FLAG-tagged BcsA as ...Details: Protein macrocomplex encompassing cytosolic (BcsRQEF) and inner-membrane (BcsAB) components. Isolated from purified and detergent-solubilised E. coli 1094 membranes using FLAG-tagged BcsA as bait. Stabilised by gentle glutaraldehyde cross-linking over a density gradient (GraFix). |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 1.4 MDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 20 mM HEPES pH 8.0 120 mM NaCl, 10%-40% glycerol, 5 mM MgCl2 10 uM AppCp 2 uM cyclic c-di-GMP cOmplete protease inhibitors |
| Staining | Type: NEGATIVE / Material: Uranyl Acetate Details: 5 micrometers of sample were spotted on glow-discharged carbon-coated copper grids. After 1 minutes the liquid was blotted off and the sample was stained by quick passage through 3 drops of ...Details: 5 micrometers of sample were spotted on glow-discharged carbon-coated copper grids. After 1 minutes the liquid was blotted off and the sample was stained by quick passage through 3 drops of 2% uranyl acetate. The sample was left for 20-30 seconds on the last drop of stain, after which the liquid was blotted off and the sample was allowed to air-dry |
| Grid | Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Details: 7mA, 10 microns |
| Details | Sample was purified from pelleted and solubilised membrane fractions of the E. coli 1094 2K7 BcsA-HA-FLAG strain using anti-FLAG affinity gel, 3xFLAG peptide elution and glycerol gradient purification coupled with gentle glutaraldehyde cross-linking (GraFix). Sample was monodisperse and subjected to thin-layer uranyl acetate staining. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Number grids imaged: 2 / Average electron dose: 12.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus min: 0.5 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)