+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | transporter | |||||||||
 Map data Map data | transporter | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transporter / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationflavin adenine dinucleotide biosynthetic process / Vitamin B2 (riboflavin) metabolism / riboflavin metabolic process / riboflavin transport / riboflavin transmembrane transporter activity / sensory perception of sound / cellular response to heat / nuclear membrane / apical plasma membrane / nucleus ...flavin adenine dinucleotide biosynthetic process / Vitamin B2 (riboflavin) metabolism / riboflavin metabolic process / riboflavin transport / riboflavin transmembrane transporter activity / sensory perception of sound / cellular response to heat / nuclear membrane / apical plasma membrane / nucleus / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.29 Å | |||||||||
 Authors Authors | Jiang DH / Wang K | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure of transporter Authors: Jiang DH / Wang K | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38623.map.gz emd_38623.map.gz | 117.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38623-v30.xml emd-38623-v30.xml emd-38623.xml emd-38623.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38623.png emd_38623.png | 37.3 KB | ||
| Filedesc metadata |  emd-38623.cif.gz emd-38623.cif.gz | 5.3 KB | ||
| Others |  emd_38623_half_map_1.map.gz emd_38623_half_map_1.map.gz emd_38623_half_map_2.map.gz emd_38623_half_map_2.map.gz | 115.9 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38623 http://ftp.pdbj.org/pub/emdb/structures/EMD-38623 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38623 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38623 | HTTPS FTP |
-Related structure data
| Related structure data |  8xsnMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38623.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38623.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | transporter | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: transporter
| File | emd_38623_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | transporter | ||||||||||||
| Projections & Slices |
| ||||||||||||
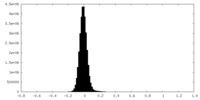
| Density Histograms |
-Half map: transporter
| File | emd_38623_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | transporter | ||||||||||||
| Projections & Slices |
| ||||||||||||
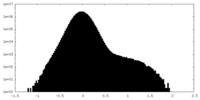
| Density Histograms |
- Sample components
Sample components
-Entire : Complex with B
| Entire | Name: Complex with B |
|---|---|
| Components |
|
-Supramolecule #1: Complex with B
| Supramolecule | Name: Complex with B / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 52, riboflavin transporter, member 3
| Macromolecule | Name: Solute carrier family 52, riboflavin transporter, member 3 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.83175 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAFLMHLLVC VFGMGSWVTI NGLWVELPLL VMELPEGWYL PSYLTVVIQL ANIGPLLVTL LHHFRPSCLS EVPIIFTLLG VGTVTCIIF AFLWNMTSWV LDGHHSIAFL VLTFFLALVD CTSSVTFLPF MSRLPTYYLT TFFVGEGLSG LLPALVALAQ G SGLTTCVN ...String: MAFLMHLLVC VFGMGSWVTI NGLWVELPLL VMELPEGWYL PSYLTVVIQL ANIGPLLVTL LHHFRPSCLS EVPIIFTLLG VGTVTCIIF AFLWNMTSWV LDGHHSIAFL VLTFFLALVD CTSSVTFLPF MSRLPTYYLT TFFVGEGLSG LLPALVALAQ G SGLTTCVN VTEISDSVPS PVPTRETDIA QGVPRALVSA LPGMEAPLSH LESRYLPAHF SPLVFFLLLS IMMACCLVAF FV LQRQPRC WEASVEDLLN DQVTLHSIRP REENDLGPAG TVDSSQGQGY LEEKAAPCCP AHLAFIYTLV AFVNALTNGM LPS VQTYSC LSYGPVAYHL AATLSIVANP LASLVSMFLP NRSLLFLGVL SVLGTCFGGY NMAMAVMSPC PLLQGHWGGE VLIV ASWVL FSGCLSYVKV MLGVVLRDLS RSALLWCGAA VQLGSLLGAL LMFPLVNVLR LFSSADFCNL HCPA UniProtKB: Solute carrier family 52, riboflavin transporter, member 3 |
-Macromolecule #2: RIBOFLAVIN
| Macromolecule | Name: RIBOFLAVIN / type: ligand / ID: 2 / Number of copies: 1 / Formula: RBF |
|---|---|
| Molecular weight | Theoretical: 376.364 Da |
| Chemical component information |  ChemComp-RBF: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)