[English] 日本語
 Yorodumi
Yorodumi- EMDB-38535: Cryo-EM structure of the ClpC1:ClpP1P2 degradation complex in Str... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the ClpC1:ClpP1P2 degradation complex in Streptomyces hawaiiensis | |||||||||
 Map data Map data | merged focus map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | protease / protein degradation / proteostasis / proteolysis / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationendopeptidase Clp / endopeptidase Clp complex / ATP-dependent peptidase activity / protein quality control for misfolded or incompletely synthesized proteins / cellular response to heat / ATPase binding / serine-type endopeptidase activity / ATP hydrolysis activity / ATP binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Streptomyces hawaiiensis (bacteria) / Streptomyces hawaiiensis (bacteria) /  | |||||||||
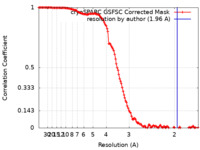
| Method | single particle reconstruction / cryo EM / Resolution: 1.96 Å | |||||||||
 Authors Authors | Xu X / Long F | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: mBio / Year: 2024 Journal: mBio / Year: 2024Title: Structural insights into the Clp protein degradation machinery. Authors: Xiaolong Xu / Yanhui Wang / Wei Huang / Danyang Li / Zixin Deng / Feng Long /  Abstract: The Clp protease system is important for maintaining proteostasis in bacteria. It consists of ClpP serine proteases and an AAA+ Clp-ATPase such as ClpC1. The hexameric ATPase ClpC1 utilizes the ...The Clp protease system is important for maintaining proteostasis in bacteria. It consists of ClpP serine proteases and an AAA+ Clp-ATPase such as ClpC1. The hexameric ATPase ClpC1 utilizes the energy of ATP binding and hydrolysis to engage, unfold, and translocate substrates into the proteolytic chamber of homo- or hetero-tetradecameric ClpP for degradation. The assembly between the hetero-tetradecameric ClpP1P2 chamber and the Clp-ATPases containing tandem ATPase domains from the same species has not been studied in depth. Here, we present cryo-EM structures of the substrate-bound ClpC1:shClpP1P2 from , and shClpP1P2 in complex with ADEP1, a natural compound produced by and known to cause over-activation and dysregulation of the ClpP proteolytic core chamber. Our structures provide detailed information on the shClpP1-shClpP2, shClpP2-ClpC1, and ADEP1-shClpP1/P2 interactions, reveal conformational transition of ClpC1 during the substrate translocation, and capture a rotational ATP hydrolysis mechanism likely dominated by the D1 ATPase activity of chaperones.IMPORTANCEThe Clp-dependent proteolysis plays an important role in bacterial homeostasis and pathogenesis. The ClpP protease system is an effective drug target for antibacterial therapy. can produce a class of potent acyldepsipeptide antibiotics such as ADEP1, which could affect the ClpP protease activity. Although hosts one of the most intricate ClpP systems in nature, very little was known about its Clp protease mechanism and the impact of ADEP molecules on ClpP. The significance of our research is in dissecting the functional mechanism of the assembled Clp degradation machinery, as well as the interaction between ADEP1 and the ClpP proteolytic chamber, by solving high-resolution structures of the substrate-bound Clp system in . The findings shed light on our understanding of the Clp-dependent proteolysis in bacteria, which will enhance the development of antimicrobial drugs targeting the Clp protease system, and help fighting against bacterial multidrug resistance. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38535.map.gz emd_38535.map.gz | 103 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38535-v30.xml emd-38535-v30.xml emd-38535.xml emd-38535.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38535_fsc.xml emd_38535_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_38535.png emd_38535.png | 203.6 KB | ||
| Masks |  emd_38535_msk_1.map emd_38535_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-38535.cif.gz emd-38535.cif.gz | 6.9 KB | ||
| Others |  emd_38535_additional_1.map.gz emd_38535_additional_1.map.gz emd_38535_half_map_1.map.gz emd_38535_half_map_1.map.gz emd_38535_half_map_2.map.gz emd_38535_half_map_2.map.gz | 204.3 MB 200.4 MB 200.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38535 http://ftp.pdbj.org/pub/emdb/structures/EMD-38535 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38535 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38535 | HTTPS FTP |
-Related structure data
| Related structure data |  8xonMC  8xn4C  8xooC  8xopC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38535.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38535.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | merged focus map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_38535_msk_1.map emd_38535_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: sharpened full map
| File | emd_38535_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened full map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_38535_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_38535_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ClpP1,ClpP2,ClpC1,casein
| Entire | Name: ClpP1,ClpP2,ClpC1,casein |
|---|---|
| Components |
|
-Supramolecule #1: ClpP1,ClpP2,ClpC1,casein
| Supramolecule | Name: ClpP1,ClpP2,ClpC1,casein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Streptomyces hawaiiensis (bacteria) Streptomyces hawaiiensis (bacteria) |
-Macromolecule #1: NDP-hexose 4-ketoreductase
| Macromolecule | Name: NDP-hexose 4-ketoreductase / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptomyces hawaiiensis (bacteria) Streptomyces hawaiiensis (bacteria) |
| Molecular weight | Theoretical: 77.312055 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MASMTGGQQM GRGSEFAEGT PSTSLVLDQF GRNLTQAARE SKLDPVIGRE KEIERVMQVL SRRTKNNPV LIGEPGVGKT AVVEGLAQAI VKGEVPETLK DKHLYTLDLG ALVAGSRYRG DFEERLKKVL KEIRTRGDII L FIDALHTL ...String: MGSSHHHHHH SSGLVPRGSH MASMTGGQQM GRGSEFAEGT PSTSLVLDQF GRNLTQAARE SKLDPVIGRE KEIERVMQVL SRRTKNNPV LIGEPGVGKT AVVEGLAQAI VKGEVPETLK DKHLYTLDLG ALVAGSRYRG DFEERLKKVL KEIRTRGDII L FIDALHTL VGAGAAEGAI DAASILKPML ARGELQTIGA TTLDEYRKHL EKDAALERRF QPIQVAEPSL PHTIEILKGL RD RYEAHHR VSITDEALVQ AATLADRYIS DRFLPDKAID LIDEAGSRMR IRRMTAPPDL REFDEKIAGV RRDKESAIDS QDA EKAASL RDKEKQLLAA KAKREKEWKA GDMDVVAEVD GELIAEVLAT ATGIPVFKLT EEESSRLLRM EDELHKRVIG QVDA VKALS KAIRRTRAGL KDPKRPGGSF IFAGPSGVGK TELSKALAEF LFGDEDALIS LDMSEFSEKH TVSRLFGSPP GYVGY EEGG QLTEKVRRKP FSVVLFDAVE KAHPDIFNSL LQILEDGRLT DSQGRVVDFK NTVIIMTTNL GTRDISKGFN LGFAAQ GDT KSNYERMKNK VSDELKQHFR PEFLNRVDDV VVFPQLSQAD ILKIVDLMID KVDERLKDRD MGIELSSSAK ELLSKKG YD PVLGARPLRR TIQREIEDSL SEKILFGELR PGHIVVVDTE GEGETKTFTF RGEE UniProtKB: NDP-hexose 4-ketoreductase |
-Macromolecule #2: casein
| Macromolecule | Name: casein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.060531 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #3: ATP-dependent Clp protease proteolytic subunit
| Macromolecule | Name: ATP-dependent Clp protease proteolytic subunit / type: protein_or_peptide / ID: 3 / Number of copies: 7 / Enantiomer: LEVO / EC number: endopeptidase Clp |
|---|---|
| Source (natural) | Organism:  Streptomyces hawaiiensis (bacteria) Streptomyces hawaiiensis (bacteria) |
| Molecular weight | Theoretical: 24.216336 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MASMTGGQQM GRGSEFGGLG DQVYNRLLNE RIIFLGQPVD DDIANKITAQ LLLLASDPEK DIYLYINSP GGSITAGMAI YDTMQYIKND VVTIAMGLAA AMGQFLLSAG TPGKRFALPN AEILIHQPSA GLAGSASDIK I HAERLLHT ...String: MGSSHHHHHH SSGLVPRGSH MASMTGGQQM GRGSEFGGLG DQVYNRLLNE RIIFLGQPVD DDIANKITAQ LLLLASDPEK DIYLYINSP GGSITAGMAI YDTMQYIKND VVTIAMGLAA AMGQFLLSAG TPGKRFALPN AEILIHQPSA GLAGSASDIK I HAERLLHT KKRMAELTSQ HTGQTIEQIT RDSDRDRWFD AFEAKEYGLI DDVMTTAAGM PGGGGTGA UniProtKB: ATP-dependent Clp protease proteolytic subunit |
-Macromolecule #4: ATP-dependent Clp protease proteolytic subunit
| Macromolecule | Name: ATP-dependent Clp protease proteolytic subunit / type: protein_or_peptide / ID: 4 / Number of copies: 7 / Enantiomer: LEVO / EC number: endopeptidase Clp |
|---|---|
| Source (natural) | Organism:  Streptomyces hawaiiensis (bacteria) Streptomyces hawaiiensis (bacteria) |
| Molecular weight | Theoretical: 22.80785 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MEYDPYAKLF EERVIFLGVQ IDDASANDVM AQLLCLESMD PDRDISVYIN SPGGSFTALT AIYDTMQYV KPDVQTVCMG QAAAAAAVLL AAGTPGKRMA LPNARVLIHQ PYSETGRGQV SDLEIAANEI LRMRSQLEDM L AKHSTTPV ...String: MGSSHHHHHH SSGLVPRGSH MEYDPYAKLF EERVIFLGVQ IDDASANDVM AQLLCLESMD PDRDISVYIN SPGGSFTALT AIYDTMQYV KPDVQTVCMG QAAAAAAVLL AAGTPGKRMA LPNARVLIHQ PYSETGRGQV SDLEIAANEI LRMRSQLEDM L AKHSTTPV EKIREDIERD KILTAEDALS YGLIDQVIST RKMDNSSLR UniProtKB: ATP-dependent Clp protease proteolytic subunit |
-Macromolecule #5: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 8 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 7 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #7: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 7 / Number of copies: 4 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 50 sec. |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model / Details: predicted models |
|---|---|
| Output model |  PDB-8xon: |
 Movie
Movie Controller
Controller








 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)