[English] 日本語
 Yorodumi
Yorodumi- EMDB-38438: Structure of human propionyl-CoA carboxylase in complex with prop... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human propionyl-CoA carboxylase in complex with propionyl-CoA (PCC-PCO) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Biotin-dependent carboxylase / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationshort-chain fatty acid catabolic process / branched-chain amino acid metabolic process / Propionyl-CoA catabolism / propionyl-CoA carboxylase / propionyl-CoA carboxylase activity / Defective HLCS causes multiple carboxylase deficiency / Biotin transport and metabolism / biotin binding / catalytic complex / Mitochondrial protein degradation ...short-chain fatty acid catabolic process / branched-chain amino acid metabolic process / Propionyl-CoA catabolism / propionyl-CoA carboxylase / propionyl-CoA carboxylase activity / Defective HLCS causes multiple carboxylase deficiency / Biotin transport and metabolism / biotin binding / catalytic complex / Mitochondrial protein degradation / fatty acid metabolic process / mitochondrial matrix / enzyme binding / mitochondrion / ATP binding / metal ion binding / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Zhou FY / Zhang YY / Zhou Q / Hu Q | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC) Authors: Zhou F / Zhang Y / Zhu Y / Zhou Q / Shi Y / Hu Q | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38438.map.gz emd_38438.map.gz | 418.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38438-v30.xml emd-38438-v30.xml emd-38438.xml emd-38438.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38438.png emd_38438.png | 31.5 KB | ||
| Filedesc metadata |  emd-38438.cif.gz emd-38438.cif.gz | 5.7 KB | ||
| Others |  emd_38438_half_map_1.map.gz emd_38438_half_map_1.map.gz emd_38438_half_map_2.map.gz emd_38438_half_map_2.map.gz | 411.9 MB 411.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38438 http://ftp.pdbj.org/pub/emdb/structures/EMD-38438 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38438 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38438 | HTTPS FTP |
-Validation report
| Summary document |  emd_38438_validation.pdf.gz emd_38438_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_38438_full_validation.pdf.gz emd_38438_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_38438_validation.xml.gz emd_38438_validation.xml.gz | 18.3 KB | Display | |
| Data in CIF |  emd_38438_validation.cif.gz emd_38438_validation.cif.gz | 21.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38438 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38438 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38438 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38438 | HTTPS FTP |
-Related structure data
| Related structure data |  8xl5MC  8xl3C  8xl4C  8xl6C  8xl7C  8xl8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38438.map.gz / Format: CCP4 / Size: 443.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38438.map.gz / Format: CCP4 / Size: 443.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0773 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_38438_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_38438_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Heterododecamer complex of human propionyl-CoA carboxylase in com...
| Entire | Name: Heterododecamer complex of human propionyl-CoA carboxylase in complex with propionyl-CoA (PCC-PCO) |
|---|---|
| Components |
|
-Supramolecule #1: Heterododecamer complex of human propionyl-CoA carboxylase in com...
| Supramolecule | Name: Heterododecamer complex of human propionyl-CoA carboxylase in complex with propionyl-CoA (PCC-PCO) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Propionyl-CoA carboxylase alpha chain, mitochondrial
| Macromolecule | Name: Propionyl-CoA carboxylase alpha chain, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: propionyl-CoA carboxylase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 80.161922 KDa |
| Sequence | String: MAGFWVGTAP LVAAGRRGRW PPQQLMLSAA LRTLKHVLYY SRQCLMVSRN LGSVGYDPNE KTFDKILVAN RGEIACRVIR TCKKMGIKT VAIHSDVDAS SVHVKMADEA VCVGPAPTSK SYLNMDAIME AIKKTRAQAV HPGYGFLSEN KEFARCLAAE D VVFIGPDT ...String: MAGFWVGTAP LVAAGRRGRW PPQQLMLSAA LRTLKHVLYY SRQCLMVSRN LGSVGYDPNE KTFDKILVAN RGEIACRVIR TCKKMGIKT VAIHSDVDAS SVHVKMADEA VCVGPAPTSK SYLNMDAIME AIKKTRAQAV HPGYGFLSEN KEFARCLAAE D VVFIGPDT HAIQAMGDKI ESKLLAKKAE VNTIPGFDGV VKDAEEAVRI AREIGYPVMI KASAGGGGKG MRIAWDDEET RD GFRLSSQ EAASSFGDDR LLIEKFIDNP RHIEIQVLGD KHGNALWLNE RECSIQRRNQ KVVEEAPSIF LDAETRRAMG EQA VALARA VKYSSAGTVE FLVDSKKNFY FLEMNTRLQV EHPVTECITG LDLVQEMIRV AKGYPLRHKQ ADIRINGWAV ECRV YAEDP YKSFGLPSIG RLSQYQEPLH LPGVRVDSGI QPGSDISIYY DPMISKLITY GSDRTEALKR MADALDNYVI RGVTH NIAL LREVIINSRF VKGDISTKFL SDVYPDGFKG HMLTKSEKNQ LLAIASSLFV AFQLRAQHFQ ENSRMPVIKP DIANWE LSV KLHDKVHTVV ASNNGSVFSV EVDGSKLNVT STWNLASPLL SVSVDGTQRT VQCLSREAGG NMSIQFLGTV YKVNILT RL AAELNKFMLE KVTEDTSSVL RSPMPGVVVA VSVKPGDAVA EGQEICVIEA MKMQNSMTAG KTGTVKSVHC QAGDTVGE G DLLVELE UniProtKB: Propionyl-CoA carboxylase alpha chain, mitochondrial |
-Macromolecule #2: Propionyl-CoA carboxylase beta chain, mitochondrial
| Macromolecule | Name: Propionyl-CoA carboxylase beta chain, mitochondrial / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO / EC number: propionyl-CoA carboxylase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 58.284488 KDa |
| Sequence | String: MAAALRVAAV GARLSVLASG LRAAVRSLCS QATSVNERIE NKRRTALLGG GQRRIDAQHK RGKLTARERI SLLLDPGSFV ESDMFVEHR CADFGMAADK NKFPGDSVVT GRGRINGRLV YVFSQDFTVF GGSLSGAHAQ KICKIMDQAI TVGAPVIGLN D SGGARIQE ...String: MAAALRVAAV GARLSVLASG LRAAVRSLCS QATSVNERIE NKRRTALLGG GQRRIDAQHK RGKLTARERI SLLLDPGSFV ESDMFVEHR CADFGMAADK NKFPGDSVVT GRGRINGRLV YVFSQDFTVF GGSLSGAHAQ KICKIMDQAI TVGAPVIGLN D SGGARIQE GVESLAGYAD IFLRNVTASG VIPQISLIMG PCAGGAVYSP ALTDFTFMVK DTSYLFITGP DVVKSVTNED VT QEELGGA KTHTTMSGVA HRAFENDVDA LCNLRDFFNY LPLSSQDPAP VRECHDPSDR LVPELDTIVP LESTKAYNMV DII HSVVDE REFFEIMPNY AKNIIVGFAR MNGRTVGIVG NQPKVASGCL DINSSVKGAR FVRFCDAFNI PLITFVDVPG FLPG TAQEY GGIIRHGAKL LYAFAEATVP KVTVITRKAY GGAYDVMSSK HLCGDTNYAW PTAEIAVMGA KGAVEIIFKG HENVE AAQA EYIEKFANPF PAAVRGFVDD IIQPSSTRAR ICCDLDVLAS KKVQRPWRKH ANIPL UniProtKB: Propionyl-CoA carboxylase beta chain, mitochondrial |
-Macromolecule #3: BIOTIN
| Macromolecule | Name: BIOTIN / type: ligand / ID: 3 / Number of copies: 6 / Formula: BTN |
|---|---|
| Molecular weight | Theoretical: 244.311 Da |
| Chemical component information | 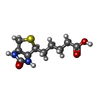 ChemComp-BTN: |
-Macromolecule #4: propionyl Coenzyme A
| Macromolecule | Name: propionyl Coenzyme A / type: ligand / ID: 4 / Number of copies: 6 / Formula: 1VU |
|---|---|
| Molecular weight | Theoretical: 823.597 Da |
| Chemical component information |  ChemComp-191: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 105293 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































