[English] 日本語
 Yorodumi
Yorodumi- EMDB-38371: Structure of Eastern Equine Encephalitis VLP in complex with the ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Eastern Equine Encephalitis VLP in complex with the receptor VLDLR LA3-5 | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | East Equine Encephalitis virus / EEEV / receptor / complex / VLDLR / glycoprotein / VIRAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationreelin receptor activity / VLDL clearance / glycoprotein transport / very-low-density lipoprotein particle receptor activity / ventral spinal cord development / Reelin signalling pathway / very-low-density lipoprotein particle binding / low-density lipoprotein particle receptor activity / very-low-density lipoprotein particle clearance / togavirin ...reelin receptor activity / VLDL clearance / glycoprotein transport / very-low-density lipoprotein particle receptor activity / ventral spinal cord development / Reelin signalling pathway / very-low-density lipoprotein particle binding / low-density lipoprotein particle receptor activity / very-low-density lipoprotein particle clearance / togavirin / reelin-mediated signaling pathway / very-low-density lipoprotein particle / positive regulation of dendrite development / cargo receptor activity / T=4 icosahedral viral capsid / lipid transport / dendrite morphogenesis / regulation of synapse assembly / apolipoprotein binding / cholesterol metabolic process / clathrin-coated pit / receptor-mediated endocytosis / VLDLR internalisation and degradation / memory / calcium-dependent protein binding / nervous system development / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / receptor complex / symbiont-mediated suppression of host gene expression / lysosomal membrane / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / calcium ion binding / symbiont entry into host cell / virion attachment to host cell / host cell nucleus / host cell plasma membrane / glutamatergic synapse / virion membrane / structural molecule activity / signal transduction / proteolysis / RNA binding / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |   Eastern equine encephalitis virus / Eastern equine encephalitis virus /  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
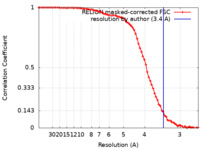
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||
 Authors Authors | Cao D / Ma B / Cao Z / Xu X / Zhang X / Xiang Y | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: The receptor VLDLR binds Eastern Equine Encephalitis virus through multiple distinct modes. Authors: Duanfang Cao / Bingting Ma / Ziyi Cao / Xiaoyu Xu / Xinzheng Zhang / Ye Xiang /  Abstract: Eastern Equine Encephalitis virus (EEEV) is an alphavirus that can cause severe diseases in infected humans. The very low-density lipoprotein receptor (VLDLR) was recently identified as a receptor of ...Eastern Equine Encephalitis virus (EEEV) is an alphavirus that can cause severe diseases in infected humans. The very low-density lipoprotein receptor (VLDLR) was recently identified as a receptor of EEEV. Herein, we performed cryo-electron microscopy structural and biochemistry studies on the specific interactions between EEEV and VLDLR. Our results show that VLDLR binds EEEV at three different sites A, B and C through its membrane-distal LDLR class A (LA) repeats. Site A is located in the cleft in between the E1-E2 heterodimers. Site B is located near the connecting β ribbon of E2 and is in proximity to site A, while site C is on the domain B of E2. The binding of VLDLR LAs to EEEV is in complex modes, including the LA1-2 and LA3-5 mediated two major modes. Disruption of the LA1-2 mediated binding significantly affect the cell attachment of EEEV. However, the mutation W132G of VLDLR impairs the binding of LA3, drives the switch of the binding modes, and significantly enhances the attachment of EEEV to the cell. The W132G variant of VLDLR could be identified in human genome and SNP sequences, implying that people with similar mutations in VLDLR may be highly susceptible to EEEV infection. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38371.map.gz emd_38371.map.gz | 38 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38371-v30.xml emd-38371-v30.xml emd-38371.xml emd-38371.xml | 22.1 KB 22.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38371_fsc.xml emd_38371_fsc.xml | 7.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_38371.png emd_38371.png | 172.2 KB | ||
| Masks |  emd_38371_msk_1.map emd_38371_msk_1.map | 40.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-38371.cif.gz emd-38371.cif.gz | 6.9 KB | ||
| Others |  emd_38371_half_map_1.map.gz emd_38371_half_map_1.map.gz emd_38371_half_map_2.map.gz emd_38371_half_map_2.map.gz | 31.2 MB 31.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38371 http://ftp.pdbj.org/pub/emdb/structures/EMD-38371 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38371 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38371 | HTTPS FTP |
-Validation report
| Summary document |  emd_38371_validation.pdf.gz emd_38371_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_38371_full_validation.pdf.gz emd_38371_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_38371_validation.xml.gz emd_38371_validation.xml.gz | 13.6 KB | Display | |
| Data in CIF |  emd_38371_validation.cif.gz emd_38371_validation.cif.gz | 19.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38371 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38371 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38371 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38371 | HTTPS FTP |
-Related structure data
| Related structure data |  8xi5MC  8xi4C  8ys2C  8ys4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38371.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38371.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.33 Å | ||||||||||||||||||||||||||||||||||||
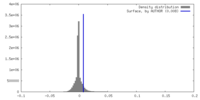
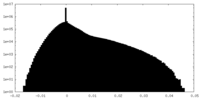
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_38371_msk_1.map emd_38371_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
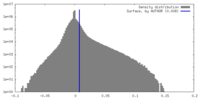
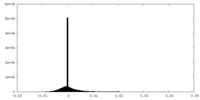
| Density Histograms |
-Half map: #2
| File | emd_38371_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
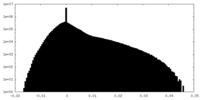
| Density Histograms |
-Half map: #1
| File | emd_38371_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : East Equine Encephalitis virus VLP in complex with its receptor V...
| Entire | Name: East Equine Encephalitis virus VLP in complex with its receptor VLDLR LA3-5 at the asymmetric unit |
|---|---|
| Components |
|
-Supramolecule #1: East Equine Encephalitis virus VLP in complex with its receptor V...
| Supramolecule | Name: East Equine Encephalitis virus VLP in complex with its receptor VLDLR LA3-5 at the asymmetric unit type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:   Eastern equine encephalitis virus / Strain: strain PE6 Eastern equine encephalitis virus / Strain: strain PE6 |
-Macromolecule #1: Spike glycoprotein E1
| Macromolecule | Name: Spike glycoprotein E1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Eastern equine encephalitis virus Eastern equine encephalitis virus |
| Molecular weight | Theoretical: 47.984246 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: YEHTAVMPNK VGIPYKALVE RPGYAPVHLQ IQLVNTRIIP STNLEYITCK YKTKVPSPVV KCCGATQCTS KPHPDYQCQV FSGVYPFMW GGAYCFCDTE NTQMSEAYVE RSEECSIDHA KAYKVHTGTV QAMVNITYGS VSWRSADVYV NGETPAKIGD A KLIIGPLS ...String: YEHTAVMPNK VGIPYKALVE RPGYAPVHLQ IQLVNTRIIP STNLEYITCK YKTKVPSPVV KCCGATQCTS KPHPDYQCQV FSGVYPFMW GGAYCFCDTE NTQMSEAYVE RSEECSIDHA KAYKVHTGTV QAMVNITYGS VSWRSADVYV NGETPAKIGD A KLIIGPLS SAWSPFDNKV VVYGHEVYNY DFPEYGTGKA GSFGDLQSRT STSNDLYANT NLKLQRPQAG IVHTPFTQVP SG FERWKKD KGAPLNDVAP FGCSIALEPL RAENCAVGSI PISIDIPDAA FTRISETPTV SDLECKITEC TYAFDFGGIA TVA YKSSKA GNCPIHSPSG VAVIKENDVT LAESGSFTFH FSTANIHPAF KLQVCTSAVT CKGDCKPPKD HIVDYPAQHT ESFT SAISA TAWSWIKVLV GGTSAFIVLG LIATAVVALV LFFHRH UniProtKB: Structural polyprotein |
-Macromolecule #2: Spike glycoprotein E2
| Macromolecule | Name: Spike glycoprotein E2 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Eastern equine encephalitis virus Eastern equine encephalitis virus |
| Molecular weight | Theoretical: 47.04702 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DLDTHFTQYK LARPYIADCP NCGHSRCDSP IAIEEVRGDA HAGVIRIQTS AMFGLKTDGV DLAYMSFMNG KTQKSIKIDN LHVRTSAPC SLVSHHGYYI LAQCPPGDTV TVGFHDGPNR HTCTVAHKVE FRPVGREKYR HPPEHGVELP CNRYTHKRAD Q GHYVEMHQ ...String: DLDTHFTQYK LARPYIADCP NCGHSRCDSP IAIEEVRGDA HAGVIRIQTS AMFGLKTDGV DLAYMSFMNG KTQKSIKIDN LHVRTSAPC SLVSHHGYYI LAQCPPGDTV TVGFHDGPNR HTCTVAHKVE FRPVGREKYR HPPEHGVELP CNRYTHKRAD Q GHYVEMHQ PGLVADHSLL SIHSAKVKIT VPSGAQVKYY CKCPDVRKGI TSSDHTTTCT DVKQCRAYLI DNKKWVYNSG RL PRGEGDT FKGKLHVPFV PVKAKCIATL APEPLVEHKH RTLILHLHPD HPTLLTTRSL GSDANPTRQW IERPTTVNFT VTG EGLEYT WGNHPPKRVW AQESGEGNPH GWPHEVVVYY YNRYPLTTII GLCTCVAIIM VSCVTSVWLL CRTRNLCITP YKLA PNAQV PILLALLCCI KPTRA UniProtKB: Structural polyprotein |
-Macromolecule #3: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 3 / Details: GenBank AAU95735.1 / Number of copies: 4 / Enantiomer: LEVO / EC number: togavirin |
|---|---|
| Source (natural) | Organism:   Eastern equine encephalitis virus Eastern equine encephalitis virus |
| Molecular weight | Theoretical: 29.178846 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFPYPTLNYP PMAPINPMAY RDPNPPRRRW RPFRPPLAAQ IEDLRRSIAS LTLKQRAPNP PAGPPANRKK PAPKPKPAQA KKKRPPPPA KKQKRKPKPG KRQRMCMKLE SDKTFPIMLN GQVNGYACVV GGRVFKPLHV EGRIDNEQLA AIKLKKASIY D LEYGDVPQ ...String: MFPYPTLNYP PMAPINPMAY RDPNPPRRRW RPFRPPLAAQ IEDLRRSIAS LTLKQRAPNP PAGPPANRKK PAPKPKPAQA KKKRPPPPA KKQKRKPKPG KRQRMCMKLE SDKTFPIMLN GQVNGYACVV GGRVFKPLHV EGRIDNEQLA AIKLKKASIY D LEYGDVPQ CMKSDTLQYT SDKPPGFYNW HHGAVQYENN RFTVPRGVGG KGDSGRPILD NKGRVVAIVL GGVNEGSRTA LS VVTWNQK GVTVKDTPEG SEPW UniProtKB: Structural polyprotein |
-Macromolecule #4: Very low-density lipoprotein receptor
| Macromolecule | Name: Very low-density lipoprotein receptor / type: protein_or_peptide / ID: 4 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 4.428696 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: PTCGAHEFQC STSSCIPISW VCDDDADCSD QSDESLEQCG R UniProtKB: Very low-density lipoprotein receptor |
-Macromolecule #5: Very low-density lipoprotein receptor
| Macromolecule | Name: Very low-density lipoprotein receptor / type: protein_or_peptide / ID: 5 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 4.532837 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RTCRIHEISC GAHSTQCIPV SWRCDGENDC DSGEDEENCG N UniProtKB: Very low-density lipoprotein receptor |
-Macromolecule #6: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 6 / Number of copies: 8 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.7 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)