+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | penton capsomer of the VZV C-capsid | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | VZV / capsid structure / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationnuclear capsid assembly / T=16 icosahedral viral capsid / viral genome packaging / viral tegument / symbiont-mediated suppression of cytoplasmic pattern recognition receptor signaling pathway / deNEDDylase activity / deubiquitinase activity / viral DNA genome replication / chromosome organization / viral penetration into host nucleus ...nuclear capsid assembly / T=16 icosahedral viral capsid / viral genome packaging / viral tegument / symbiont-mediated suppression of cytoplasmic pattern recognition receptor signaling pathway / deNEDDylase activity / deubiquitinase activity / viral DNA genome replication / chromosome organization / viral penetration into host nucleus / viral capsid / host cell / symbiont-mediated perturbation of host ubiquitin-like protein modification / host cell cytoplasm / ubiquitinyl hydrolase 1 / cysteine-type deubiquitinase activity / Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases / symbiont entry into host cell / host cell nucleus / structural molecule activity / proteolysis Similarity search - Function | |||||||||
| Biological species |  Human alphaherpesvirus 3 (Varicella-zoster virus) Human alphaherpesvirus 3 (Varicella-zoster virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Nan W / Lei C / Xiangxi W | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Hlife / Year: 2024 Journal: Hlife / Year: 2024Title: Insights into varicella-zoster virus assembly from the B- and C-capsid at near-atomic resolution structures Authors: Cao L / Wang N / Lv Z / Chen W / Chen Z / Song L / Sha X / Wang G / Hu Y / Lian X / Cui G / Fan J / Quan Y / Liu H / Hou H / Wang X | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38190.map.gz emd_38190.map.gz | 49 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38190-v30.xml emd-38190-v30.xml emd-38190.xml emd-38190.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38190.png emd_38190.png | 96.8 KB | ||
| Filedesc metadata |  emd-38190.cif.gz emd-38190.cif.gz | 7.2 KB | ||
| Others |  emd_38190_half_map_1.map.gz emd_38190_half_map_1.map.gz emd_38190_half_map_2.map.gz emd_38190_half_map_2.map.gz | 40.7 MB 40.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38190 http://ftp.pdbj.org/pub/emdb/structures/EMD-38190 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38190 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38190 | HTTPS FTP |
-Related structure data
| Related structure data |  8xa0MC  8x9wC  8x9xC  8x9yC  8x9zC  8xa1C  8xa2C  8xa3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38190.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38190.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||
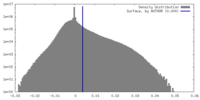
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_38190_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_38190_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human alphaherpesvirus 3
| Entire | Name:  Human alphaherpesvirus 3 (Varicella-zoster virus) Human alphaherpesvirus 3 (Varicella-zoster virus) |
|---|---|
| Components |
|
-Supramolecule #1: Human alphaherpesvirus 3
| Supramolecule | Name: Human alphaherpesvirus 3 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10335 / Sci species name: Human alphaherpesvirus 3 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: No |
|---|
-Macromolecule #1: Large tegument protein deneddylase
| Macromolecule | Name: Large tegument protein deneddylase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: ubiquitinyl hydrolase 1 |
|---|---|
| Source (natural) | Organism:  Human alphaherpesvirus 3 (Varicella-zoster virus) Human alphaherpesvirus 3 (Varicella-zoster virus) |
| Molecular weight | Theoretical: 5.50051 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QRTGRSALAV LIRACYRLQQ QLQRTRRALL HHSDAVLTSL HHVRMLL UniProtKB: Large tegument protein deneddylase |
-Macromolecule #2: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 2 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human alphaherpesvirus 3 (Varicella-zoster virus) Human alphaherpesvirus 3 (Varicella-zoster virus) |
| Molecular weight | Theoretical: 152.365312 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: IPAGIIPTGN VLSTIEVCIF FCIFDFFKQI RSDDNSLYSA QFDILLGTYC NTLNFVRFLE LGLSVACICT KFPELAYVRD GVIQFEVQQ PMIARDGPHP VDQPVHNYMV KRIHKRSLSA AFAIASEALS LLSNTYVAAT EIDSSLRIRA IQQMARNLRT V SDSFERGT ...String: IPAGIIPTGN VLSTIEVCIF FCIFDFFKQI RSDDNSLYSA QFDILLGTYC NTLNFVRFLE LGLSVACICT KFPELAYVRD GVIQFEVQQ PMIARDGPHP VDQPVHNYMV KRIHKRSLSA AFAIASEALS LLSNTYVAAT EIDSSLRIRA IQQMARNLRT V SDSFERGT ADQLLGVLLE KAPPLSLLSP INKFQPEGHL NRVARAALLS DLKRRVCADM FFMTRHAREP RLISAYLSDM VS CTQPSVM VSRITHTNTR GRQVDGVLVT TATLKRQLLQ GILQIDDTAA DVPVTYGEMV LQGTNLVTAL VMGKAVRGMD DVA RHLLDI TDPNTLNIPS IPPQSNSDST TAGLPVNARV PADLVIVGDK LVFLEALERR VYQATRVAYP LIGNIDITFI MPMG VFQAN SMDRYTRHAG DFSTVSEQDP RQFPPQGIFF YNKDGILTQL TLRDAMGTIC HSSLLDVEAT LVALRQQHLD RQCYF GVYV AEGTEDTLDV QMGRFMETWA DMMPHHPHWV NEHLTILQFI APSNPRLRFE LNPAFDFFVA PGDVDLPGPQ RPPEAM PTV NATLRIINGN IPVPLCPISF RDCRGTQLGL GRHTMTPATI KAVKDTFEDR AYPTIFYMLE AVIHGNERNF CALLRLL TQ CIRGYWEQSH RVAFVNNFHM LMYITTYLGN GELPEVCINI YRDLLQHVRA LRQTITDFTI QGEGHNGETS EALNNILT D DTFIAPILWD CDALIYRDEA ARDRLPAIRV SGRNGYQALH FVDMAGHNFQ RRDNVLIHGR PVRGDTGQAI PITPHHDRE WGILSKIYYY IVIPAFSRGS CCTMGVRYDR LYPALQAVIV PEIPADEEAP TTPEDPRHPL HAHQLVPNSL NVYFHNAHLT VDGDALLTL QELMGDMAER TTAILVSSAP DAGAATATTR NMRIYDGALY HGLIMMAYQA YDETIATGTF FYPVPVNPLF A CPEHLASL RGMTNARRVL AKMVPPIPPF LGANHHATIR QPVAYHVTHS KSDFNTLTYS LLGGYFKFTP ISLTHQLRTG FH PGIAFTV VRQDRFATEQ LLYAERASES YFVGQIQVHH HDAIGGVNFT LTQPRAHVDL GVGYTAVCAT AALRCPLTDM GNT AQNLFF SRGGVPMLHD NVTESLRRIT ASGGRLNPTE PLPIFGGLRP ATSAGIARGQ ASVCEFVAMP VSTDLQYFRT ACNP RGRAS GMLYMGDRDA DIEAIMFDHT QSDVAYTDRA TLNPWASQKH SYGDRLYNGT YNLTGASPIY SPCFKFFTPA EVNTN CNTL DRLLMEAKAV ASQSSTDTEY QFKRPPGSTE MTQDPCGLFQ EAYPPLCSSD AAMLRTAHAG ETGADEVHLA QYLIRD ASP LRGCLPL UniProtKB: Major capsid protein |
-Macromolecule #3: Capsid vertex component 1
| Macromolecule | Name: Capsid vertex component 1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human alphaherpesvirus 3 (Varicella-zoster virus) Human alphaherpesvirus 3 (Varicella-zoster virus) |
| Molecular weight | Theoretical: 73.998547 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MNAHLANEVQ YDLGHARVGP SSLVHVIISS ECLAAAGIPL AALMRGRPGL GTAANFQVEI QTRAHATGDC TPWCTAFAAY VPADAVGEL LAPVVPAHPG LLPRASSAGG LFVSLPVVCD AQGVYDPYAV AALRLAWGSG ASCARVILFS YDELVPPNTR Y AADSTRIM ...String: MNAHLANEVQ YDLGHARVGP SSLVHVIISS ECLAAAGIPL AALMRGRPGL GTAANFQVEI QTRAHATGDC TPWCTAFAAY VPADAVGEL LAPVVPAHPG LLPRASSAGG LFVSLPVVCD AQGVYDPYAV AALRLAWGSG ASCARVILFS YDELVPPNTR Y AADSTRIM RVCRHLCRYV ALLGAAAPPA AKEAAAHLSM GLGESASPRP QPLARPHAGA PADPPIVGAS DPPISPEEQL TA PGGDTTA AQDVSIAQEN EEILALVQRA VQDVTRRHPV RARTGRAACG VASGLRQGAL VHQAVSGGAM GAADADAVLA GLE PPGGGR FVAPAPHGPG GEDILNDVLT LTPGTAKPRS LVEWLDRGWE ALAGGDRPDW LWSRRSISVV LRHHYGTKQR FVVV SYENS VAWGGRRARP PLLSSALATA LTEACAAERV VRPHQLSPAG QAELLLRFPA LEVPLRHPRP VLPPFDIAAE VAFTA RIHL ACLRALGQAI RAALQGGPRI SQRLRYDFGP DQRAWLGEVT RRFPILLENL MRAVEGTAPD AFFHTAYALA VLAHLG GRG GRGRRVVPLG DDLPARFADS DGHYVFDYYS TSGDTLRLNN RPIAVAMDGD VSKREQSKCR FMEAVPSTAP RRVCEQY LP GESYAYLCLG FNRRLCGIVV FPGGFAFTIN IAAYLSLSDP VARAAVLRFC RKVS UniProtKB: Capsid vertex component 1 |
-Macromolecule #4: Capsid vertex component 2
| Macromolecule | Name: Capsid vertex component 2 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human alphaherpesvirus 3 (Varicella-zoster virus) Human alphaherpesvirus 3 (Varicella-zoster virus) |
| Molecular weight | Theoretical: 10.890343 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDPYCPFDAL DVWEHRRFIV ADSRNFITPE FPRDFWMSPV FNLPRETAAE QVVVLQAQRT AAAAALENAA MQAAELPVDI ERRLRPIER NVHEI UniProtKB: Capsid vertex component 2 |
-Macromolecule #5: Tri2A
| Macromolecule | Name: Tri2A / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human alphaherpesvirus 3 (Varicella-zoster virus) Human alphaherpesvirus 3 (Varicella-zoster virus) |
| Molecular weight | Theoretical: 27.729641 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AMPFEIEVLL PGELSPAETS ALQKCEGKII TFSTLRHRAS LVDIALSSYY INGAPPDTLS LLEAYRMRFA AVITRVIPGK LLAHAIGVG TPTPGLFIQN TSPVDLCNGD YICLLPPVYG SADSIRLDSV GLEIVFPLTI PQTLMREIIA KVVARAVEDL N LMFSINEG ...String: AMPFEIEVLL PGELSPAETS ALQKCEGKII TFSTLRHRAS LVDIALSSYY INGAPPDTLS LLEAYRMRFA AVITRVIPGK LLAHAIGVG TPTPGLFIQN TSPVDLCNGD YICLLPPVYG SADSIRLDSV GLEIVFPLTI PQTLMREIIA KVVARAVEDL N LMFSINEG CLLILALIPR LLALLIPRLL ALVTREAAQL IHPEAPMLML PIYETISSWI STSSRLGDTL GTRAILRVCV FD GPSTVHP GDRTAVIQV |
-Macromolecule #6: Tri2B
| Macromolecule | Name: Tri2B / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human alphaherpesvirus 3 (Varicella-zoster virus) Human alphaherpesvirus 3 (Varicella-zoster virus) |
| Molecular weight | Theoretical: 28.721295 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AMPFEIEVLL PGEISPAETS ALQKCEGKII TFSTLRHRAS LVDIALSSYY INGAPPDTLS LLEAYRMRFA AVITRVIPGK LLAHAIGVG TPTPGLFIQN TSPVDLCNGD YICLLPPVFG SADEIRLDSV GLEIVFPLTI PQTLMREIIA KVVARAVERT A ADVICYNG ...String: AMPFEIEVLL PGEISPAETS ALQKCEGKII TFSTLRHRAS LVDIALSSYY INGAPPDTLS LLEAYRMRFA AVITRVIPGK LLAHAIGVG TPTPGLFIQN TSPVDLCNGD YICLLPPVFG SADEIRLDSV GLEIVFPLTI PQTLMREIIA KVVARAVERT A ADVICYNG RRYELETNLQ HRDGSDAAIR TLVLNLMFSI NEGTTLILTL ITRLLRFPIY EAISSWISTS SRLGDTLGTR AI LRVCVFD GPSTVHPGDR TAVIQV |
-Macromolecule #7: Tri1
| Macromolecule | Name: Tri1 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human alphaherpesvirus 3 (Varicella-zoster virus) Human alphaherpesvirus 3 (Varicella-zoster virus) |
| Molecular weight | Theoretical: 32.26891 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: FKSTTQLIQQ VSLTDFFRPD IEHAGSTVLI LRHPTDLPAL ARHRAPPGRQ TERLAEAWGQ LLEASRAYVT SLSFIAACRA EEYTDKQAA EANRTAIVSA YGCSRMGARL IRFSECLRAM VQCHVFPHRF ISFFGSLLEY TIQDNLCNIT AVAKGPQEAA R TDKTSTRR ...String: FKSTTQLIQQ VSLTDFFRPD IEHAGSTVLI LRHPTDLPAL ARHRAPPGRQ TERLAEAWGQ LLEASRAYVT SLSFIAACRA EEYTDKQAA EANRTAIVSA YGCSRMGARL IRFSECLRAM VQCHVFPHRF ISFFGSLLEY TIQDNLCNIT AVAKGPQEAA R TDKTSTRR VTANIPACVF WDVDKDLHLS ADGLKHVFLV FVYTQRRQRE GVRLHLALSQ LNEQCFGRGI GFLLGARICM YA AYTLIGT IPSESVRYTR RMERFGGYNV PTIWLEGVVW GGTNTWNEC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.3 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 BASE (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 417864 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)