[English] 日本語
 Yorodumi
Yorodumi- EMDB-38161: Structure of human TRPV1 in complex with antagonist --protein pur... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human TRPV1 in complex with antagonist --protein purified without CHS | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationchemosensory behavior / response to capsazepine / sensory perception of mechanical stimulus / peptide secretion / excitatory extracellular ligand-gated monoatomic ion channel activity / temperature-gated ion channel activity / detection of chemical stimulus involved in sensory perception of pain / smooth muscle contraction involved in micturition / fever generation / detection of temperature stimulus involved in thermoception ...chemosensory behavior / response to capsazepine / sensory perception of mechanical stimulus / peptide secretion / excitatory extracellular ligand-gated monoatomic ion channel activity / temperature-gated ion channel activity / detection of chemical stimulus involved in sensory perception of pain / smooth muscle contraction involved in micturition / fever generation / detection of temperature stimulus involved in thermoception / thermoception / cellular response to acidic pH / dendritic spine membrane / TRP channels / diet induced thermogenesis / cellular response to alkaloid / cellular response to ATP / detection of temperature stimulus involved in sensory perception of pain / intracellularly gated calcium channel activity / behavioral response to pain / calcium ion import across plasma membrane / voltage-gated calcium channel activity / extracellular ligand-gated monoatomic ion channel activity / phosphatidylinositol binding / phosphoprotein binding / GABA-ergic synapse / calcium ion transmembrane transport / calcium channel activity / lipid metabolic process / transmembrane signaling receptor activity / sensory perception of taste / cellular response to heat / protein homotetramerization / postsynaptic membrane / calmodulin binding / cell surface receptor signaling pathway / negative regulation of transcription by RNA polymerase II / ATP binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.55 Å | |||||||||
 Authors Authors | Fan J / Lei X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis of TRPV1 inhibition by SAF312 and cholesterol. Authors: Junping Fan / Han Ke / Jing Lei / Jin Wang / Makoto Tominaga / Xiaoguang Lei /   Abstract: Transient Receptor Potential Vanilloid 1 (TRPV1) plays a central role in pain sensation and is thus an attractive pharmacological drug target. SAF312 is a potent, selective, and non-competitive ...Transient Receptor Potential Vanilloid 1 (TRPV1) plays a central role in pain sensation and is thus an attractive pharmacological drug target. SAF312 is a potent, selective, and non-competitive antagonist of TRPV1 and shows promising potential in treating ocular surface pain. However, the precise mechanism by which SAF312 inhibits TRPV1 remains poorly understood. Here, we present the cryo-EM structure of human TRPV1 in complex with SAF312, elucidating the structural foundation of its antagonistic effects on TRPV1. SAF312 binds to the vanilloid binding pocket, preventing conformational changes in S4 and S5 helices, which are essential for channel gating. Unexpectedly, a putative cholesterol was found to contribute to SAF312's inhibition. Complemented by mutagenesis experiments and molecular dynamics simulations, our research offers substantial mechanistic insights into the regulation of TRPV1 by SAF312, highlighting the interplay between the antagonist and cholesterol in modulating TRPV1 function. This work not only expands our understanding of TRPV1 inhibition by SAF312 but also lays the groundwork for further developments in the design and optimization of TRPV1-related therapies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38161.map.gz emd_38161.map.gz | 117.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38161-v30.xml emd-38161-v30.xml emd-38161.xml emd-38161.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38161.png emd_38161.png | 63.9 KB | ||
| Filedesc metadata |  emd-38161.cif.gz emd-38161.cif.gz | 6.7 KB | ||
| Others |  emd_38161_half_map_1.map.gz emd_38161_half_map_1.map.gz emd_38161_half_map_2.map.gz emd_38161_half_map_2.map.gz | 115.2 MB 115.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38161 http://ftp.pdbj.org/pub/emdb/structures/EMD-38161 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38161 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38161 | HTTPS FTP |
-Validation report
| Summary document |  emd_38161_validation.pdf.gz emd_38161_validation.pdf.gz | 843.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_38161_full_validation.pdf.gz emd_38161_full_validation.pdf.gz | 843.3 KB | Display | |
| Data in XML |  emd_38161_validation.xml.gz emd_38161_validation.xml.gz | 14 KB | Display | |
| Data in CIF |  emd_38161_validation.cif.gz emd_38161_validation.cif.gz | 16.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38161 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38161 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38161 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38161 | HTTPS FTP |
-Related structure data
| Related structure data |  8x94MC  8jqrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38161.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38161.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_38161_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
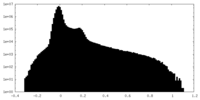
| Projections & Slices |
| ||||||||||||
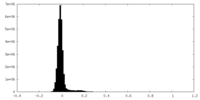
| Density Histograms |
-Half map: #2
| File | emd_38161_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
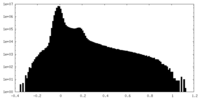
| Projections & Slices |
| ||||||||||||
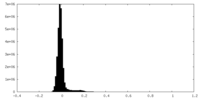
| Density Histograms |
- Sample components
Sample components
-Entire : TRPV1
| Entire | Name: TRPV1 |
|---|---|
| Components |
|
-Supramolecule #1: TRPV1
| Supramolecule | Name: TRPV1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 360 KDa |
-Macromolecule #1: Transient receptor potential cation channel subfamily V member 1,...
| Macromolecule | Name: Transient receptor potential cation channel subfamily V member 1,Green fluorescent protein type: protein_or_peptide / ID: 1 Details: The section (840-842) is the cloning site.The domain (843-850) is PreScission Site.The domain (851-1084) is corresponding to this sfGFP (462-695 amino acids, GenBank: ALP48449.1). The domain ...Details: The section (840-842) is the cloning site.The domain (843-850) is PreScission Site.The domain (851-1084) is corresponding to this sfGFP (462-695 amino acids, GenBank: ALP48449.1). The domain (1085-1112) is the expression Tag. Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 125.297734 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKKWSSTDLG AAADPLQKDT CPDPLDGDPN SRPPPAKPQL STAKSRTRLF GKGDSEEAFP VDCPHEEGEL DSCPTITVSP VITIQRPGD GPTGARLLSQ DSVAASTEKT LRLYDRRSIF EAVAQNNCQD LESLLLFLQK SKKHLTDNEF KDPETGKTCL L KAMLNLHD ...String: MKKWSSTDLG AAADPLQKDT CPDPLDGDPN SRPPPAKPQL STAKSRTRLF GKGDSEEAFP VDCPHEEGEL DSCPTITVSP VITIQRPGD GPTGARLLSQ DSVAASTEKT LRLYDRRSIF EAVAQNNCQD LESLLLFLQK SKKHLTDNEF KDPETGKTCL L KAMLNLHD GQNTTIPLLL EIARQTDSLK ELVNASYTDS YYKGQTALHI AIERRNMALV TLLVENGADV QAAAHGDFFK KT KGRPGFY FGELPLSLAA CTNQLGIVKF LLQNSWQTAD ISARDSVGNT VLHALVEVAD NTADNTKFVT SMYNEILMLG AKL HPTLKL EELTNKKGMT PLALAAGTGK IGVLAYILQR EIQEPECRHL SRKFTEWAYG PVHSSLYDLS CIDTCEKNSV LEVI AYSSS ETPNRHDMLL VEPLNRLLQD KWDRFVKRIF YFNFLVYCLY MIIFTMAAYY RPVDGLPPFK MEKTGDYFRV TGEIL SVLG GVYFFFRGIQ YFLQRRPSMK TLFVDSYSEM LFFLQSLFML ATVVLYFSHL KEYVASMVFS LALGWTNMLY YTRGFQ QMG IYAVMIEKMI LRDLCRFMFV YIVFLFGFST AVVTLIEDGK NDSLPSESTS HRWRGPACRP PDSSYNSLYS TCLELFK FT IGMGDLEFTE NYDFKAVFII LLLAYVILTY ILLLNMLIAL MGETVNKIAQ ESKNIWKLQR AITILDTEKS FLKCMRKA F RSGKLLQVGY TPDGKDDYRW CFRVDEVNWT TWNTNVGIIN EDPGNCEGVK RTLSFSLRSS RVSGRHWKNF ALVPLLREA SARDRQSAQP EEVYLRQFSG SLKPEDAEVF KSPAASGEKA AALEVLFQGP SKGEELFTGV VPILVELDGD VNGHKFSVRG EGEGDATNG KLTLKFICTT GKLPVPWPTL VTTLTYGVQC FSRYPDHMKR HDFFKSAMPE GYVQERTISF KDDGTYKTRA E VKFEGDTL VNRIELKGID FKEDGNILGH KLEYNFNSHN VYITADKQKN GIKANFKIRH NVEDGSVQLA DHYQQNTPIG DG PVLLPDN HYLSTQSVLS KDPNEKRDHM VLLEFVTAAG ITHGMDEWSH PQFEKGGGSG GGSGGSAWSH PQFEK UniProtKB: Transient receptor potential cation channel subfamily V member 1 |
-Macromolecule #2: 4-(7-Hydroxy-2-isopropyl-4-oxoquinazolin-3(4H)-yl)benzonitrile
| Macromolecule | Name: 4-(7-Hydroxy-2-isopropyl-4-oxoquinazolin-3(4H)-yl)benzonitrile type: ligand / ID: 2 / Number of copies: 4 / Formula: EZI |
|---|---|
| Molecular weight | Theoretical: 305.331 Da |
-Macromolecule #3: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 3 / Number of copies: 4 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)