+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human DRA (SLC26A3) bound with tenidap | |||||||||
 Map data Map data | full map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DRA / SLC26A3 / tenidap / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDefective SLC26A3 causes congenital secretory chloride diarrhea 1 (DIAR1) / Inorganic anion exchange by SLC26 transporters / sulfate transmembrane transporter activity / oxalate transmembrane transporter activity / sulfate transmembrane transport / secondary active sulfate transmembrane transporter activity / intracellular pH elevation / chloride:bicarbonate antiporter activity / solute:inorganic anion antiporter activity / bicarbonate transmembrane transporter activity ...Defective SLC26A3 causes congenital secretory chloride diarrhea 1 (DIAR1) / Inorganic anion exchange by SLC26 transporters / sulfate transmembrane transporter activity / oxalate transmembrane transporter activity / sulfate transmembrane transport / secondary active sulfate transmembrane transporter activity / intracellular pH elevation / chloride:bicarbonate antiporter activity / solute:inorganic anion antiporter activity / bicarbonate transmembrane transporter activity / monoatomic anion transport / membrane hyperpolarization / chloride transmembrane transporter activity / sperm capacitation / monoatomic ion transport / sperm midpiece / chloride transmembrane transport / cellular response to cAMP / brush border membrane / apical plasma membrane / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.37 Å | |||||||||
 Authors Authors | Yang X / Zhang Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of human DRA (SLC26A3) bound with tenidap Authors: Yang X / Zhang Y | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38000.map.gz emd_38000.map.gz | 97.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38000-v30.xml emd-38000-v30.xml emd-38000.xml emd-38000.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38000_fsc.xml emd_38000_fsc.xml | 9.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_38000.png emd_38000.png | 78.4 KB | ||
| Masks |  emd_38000_msk_1.map emd_38000_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-38000.cif.gz emd-38000.cif.gz | 6.1 KB | ||
| Others |  emd_38000_half_map_1.map.gz emd_38000_half_map_1.map.gz emd_38000_half_map_2.map.gz emd_38000_half_map_2.map.gz | 95.4 MB 95.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38000 http://ftp.pdbj.org/pub/emdb/structures/EMD-38000 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38000 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38000 | HTTPS FTP |
-Related structure data
| Related structure data |  8x1sMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38000.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38000.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.932 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_38000_msk_1.map emd_38000_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half B map
| File | emd_38000_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half B map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half A map
| File | emd_38000_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half A map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : DRA
| Entire | Name: DRA |
|---|---|
| Components |
|
-Supramolecule #1: DRA
| Supramolecule | Name: DRA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Chloride anion exchanger
| Macromolecule | Name: Chloride anion exchanger / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 85.877844 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDDDDKG TMIEPFGNQY IVARPVYSTN AFEENHKKTG RHHKTFLDHL KVCCSCSPQK AKRIVLSLFP IASWLPAYRL KEWLLSDIV SGISTGIVAV LQGLAFALLV DIPPVYGLYA SFFPAIIYLF FGTSRHISVG PFPILSMMVG LAVSGAVSKA V PDRNATTL ...String: MDYKDDDDKG TMIEPFGNQY IVARPVYSTN AFEENHKKTG RHHKTFLDHL KVCCSCSPQK AKRIVLSLFP IASWLPAYRL KEWLLSDIV SGISTGIVAV LQGLAFALLV DIPPVYGLYA SFFPAIIYLF FGTSRHISVG PFPILSMMVG LAVSGAVSKA V PDRNATTL GLPNNSNNSS LLDDERVRVA AAASVTVLSG IIQLAFGILR IGFVVIYLSE SLISGFTTAA AVHVLVSQLK FI FQLTVPS HTDPVSIFKV LYSVFSQIEK TNIADLVTAL IVLLVVSIVK EINQRFKDKL PVPIPIEFIM TVIAAGVSYG CDF KNRFKV AVVGDMNPGF QPPITPDVET FQNTVGDCFG IAMVAFAVAF SVASVYSLKY DYPLDGNQEL IALGLGNIVC GVFR GFAGS TALSRSAVQE STGGKTQIAG LIGAIIVLIV VLAIGFLLAP LQKSVLAALA LGNLKGMLMQ FAEIGRLWRK DKYDC LIWI MTFIFTIVLG LGLGLAASVA FQLLTIVFRT QFPKCSTLAN IGRTNIYKNK KDYYDMYEPE GVKIFRCPSP IYFANI GFF RRKLIDAVGF SPLRILRKRN KALRKIRKLQ KQGLLQVTPK GFICTVDTIK DSDEELDNNQ IEVLDQPINT TDLPFHI DW NDDLPLNIEV PKISLHSLIL DFSAVSFLDV SSVRGLKSIL QEFIRIKVDV YIVGTDDDFI EKLNRYEFFD GEVKSSIF F LTIHDAVLHI LMKKDYSTSK FNPSQEKDGK IDFTINTNGG LRNRVYEVPV ETKF UniProtKB: Chloride anion exchanger |
-Macromolecule #2: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #3: 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 3 / Number of copies: 2 / Formula: LPE |
|---|---|
| Molecular weight | Theoretical: 510.708 Da |
| Chemical component information |  ChemComp-LPE: |
-Macromolecule #4: 5-chloranyl-2-oxidanyl-3-thiophen-2-ylcarbonyl-indole-1-carboxamide
| Macromolecule | Name: 5-chloranyl-2-oxidanyl-3-thiophen-2-ylcarbonyl-indole-1-carboxamide type: ligand / ID: 4 / Number of copies: 2 / Formula: I2U |
|---|---|
| Molecular weight | Theoretical: 320.751 Da |
| Chemical component information | 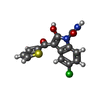 ChemComp-I2U: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 12 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Number grids imaged: 1 / Number real images: 2838 / Average exposure time: 3.89 sec. / Average electron dose: 49.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































