[English] 日本語
 Yorodumi
Yorodumi- EMDB-37958: Cryo-EM map for Mumps Virus L Protein Bound by Phosphoprotein Tet... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map for Mumps Virus L Protein Bound by Phosphoprotein Tetramer (Focused map for CD-MTase-CTD) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mumps virus polymerase complex / RNA-dependent RNA synthesis / Large protein / phosphoprotein. / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationNNS virus cap methyltransferase / GDP polyribonucleotidyltransferase / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / virion component / host cell cytoplasm / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / hydrolase activity / RNA-directed RNA polymerase / RNA-directed RNA polymerase activity / ATP binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
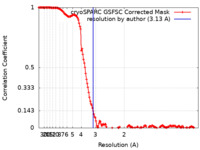
| Method | single particle reconstruction / cryo EM / Resolution: 3.13 Å | |||||||||
 Authors Authors | Li TH / Shen QT | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structures of the mumps virus polymerase complex via cryo-electron microscopy. Authors: Tianhao Li / Mingdong Liu / Zhanxi Gu / Xin Su / Yunhui Liu / Jinzhong Lin / Yu Zhang / Qing-Tao Shen /  Abstract: The viral polymerase complex, comprising the large protein (L) and phosphoprotein (P), is crucial for both genome replication and transcription in non-segmented negative-strand RNA viruses (nsNSVs), ...The viral polymerase complex, comprising the large protein (L) and phosphoprotein (P), is crucial for both genome replication and transcription in non-segmented negative-strand RNA viruses (nsNSVs), while structures corresponding to these activities remain obscure. Here, we resolved two L-P complex conformations from the mumps virus (MuV), a typical member of nsNSVs, via cryogenic-electron microscopy. One conformation presents all five domains of L forming a continuous RNA tunnel to the methyltransferase domain (MTase), preferably as a transcription state. The other conformation has the appendage averaged out, which is inaccessible to MTase. In both conformations, parallel P tetramers are revealed around MuV L, which, together with structures of other nsNSVs, demonstrates the diverse origins of the L-binding X domain of P. Our study links varying structures of nsNSV polymerase complexes with genome replication and transcription and points to a sliding model for polymerase complexes to advance along the RNA templates. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37958.map.gz emd_37958.map.gz | 305.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37958-v30.xml emd-37958-v30.xml emd-37958.xml emd-37958.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37958_fsc.xml emd_37958_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_37958.png emd_37958.png | 45.5 KB | ||
| Filedesc metadata |  emd-37958.cif.gz emd-37958.cif.gz | 4.3 KB | ||
| Others |  emd_37958_additional_1.map.gz emd_37958_additional_1.map.gz emd_37958_half_map_1.map.gz emd_37958_half_map_1.map.gz emd_37958_half_map_2.map.gz emd_37958_half_map_2.map.gz | 287.7 MB 301.6 MB 301.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37958 http://ftp.pdbj.org/pub/emdb/structures/EMD-37958 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37958 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37958 | HTTPS FTP |
-Validation report
| Summary document |  emd_37958_validation.pdf.gz emd_37958_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37958_full_validation.pdf.gz emd_37958_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_37958_validation.xml.gz emd_37958_validation.xml.gz | 23.3 KB | Display | |
| Data in CIF |  emd_37958_validation.cif.gz emd_37958_validation.cif.gz | 30 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37958 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37958 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37958 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37958 | HTTPS FTP |
-Related structure data
| Related structure data |  8yxlMC  8izlC  8x01C  8yxmC  8yxoC  8yxpC  8yxrC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37958.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37958.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.53 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Post-processing in deepEMhancer
| File | emd_37958_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processing in deepEMhancer | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_37958_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_37958_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The MuV polymerase complex of RNA-directed RNA polymerase L with ...
| Entire | Name: The MuV polymerase complex of RNA-directed RNA polymerase L with tetrameric phosphoproteins |
|---|---|
| Components |
|
-Supramolecule #1: The MuV polymerase complex of RNA-directed RNA polymerase L with ...
| Supramolecule | Name: The MuV polymerase complex of RNA-directed RNA polymerase L with tetrameric phosphoproteins type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: The MuV polymerase complex expressed in Sf9 cells and purified by affinity chromatography and size-exlusive chromatography sequentially. |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 2.75 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)