[English] 日本語
 Yorodumi
Yorodumi- EMDB-3786: Cryo-electron tomography averaged map of microtubule doublet from... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3786 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron tomography averaged map of microtubule doublet from p23 mutant Chlamydomonas axoneme | |||||||||
 Map data Map data | Subaverage after subtomogram classification of Chlamydomonas pf23 cilia, based on outer dynein arm structure | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 50.0 Å | |||||||||
 Authors Authors | Sale WS / Ishikawa T / Yamamoto R / Obbineni JM / Alford LM / Hwang J / Ide T / Owa M / Kon T / Inaba K ...Sale WS / Ishikawa T / Yamamoto R / Obbineni JM / Alford LM / Hwang J / Ide T / Owa M / Kon T / Inaba K / Noliyanda J / King SM / Dutcher S | |||||||||
 Citation Citation |  Journal: PLoS Genet / Year: 2017 Journal: PLoS Genet / Year: 2017Title: Chlamydomonas DYX1C1/PF23 is essential for axonemal assembly and proper morphology of inner dynein arms. Authors: Ryosuke Yamamoto / Jagan M Obbineni / Lea M Alford / Takahiro Ide / Mikito Owa / Juyeon Hwang / Takahide Kon / Kazuo Inaba / Noliyanda James / Stephen M King / Takashi Ishikawa / Winfield S ...Authors: Ryosuke Yamamoto / Jagan M Obbineni / Lea M Alford / Takahiro Ide / Mikito Owa / Juyeon Hwang / Takahide Kon / Kazuo Inaba / Noliyanda James / Stephen M King / Takashi Ishikawa / Winfield S Sale / Susan K Dutcher /    Abstract: Cytoplasmic assembly of ciliary dyneins, a process known as preassembly, requires numerous non-dynein proteins, but the identities and functions of these proteins are not fully elucidated. Here, we ...Cytoplasmic assembly of ciliary dyneins, a process known as preassembly, requires numerous non-dynein proteins, but the identities and functions of these proteins are not fully elucidated. Here, we show that the classical Chlamydomonas motility mutant pf23 is defective in the Chlamydomonas homolog of DYX1C1. The pf23 mutant has a 494 bp deletion in the DYX1C1 gene and expresses a shorter DYX1C1 protein in the cytoplasm. Structural analyses, using cryo-ET, reveal that pf23 axonemes lack most of the inner dynein arms. Spectral counting confirms that DYX1C1 is essential for the assembly of the majority of ciliary inner dynein arms (IDA) as well as a fraction of the outer dynein arms (ODA). A C-terminal truncation of DYX1C1 shows a reduction in a subset of these ciliary IDAs. Sucrose gradients of cytoplasmic extracts show that preassembled ciliary dyneins are reduced compared to wild-type, which suggests an important role in dynein complex stability. The role of PF23/DYX1C1 remains unknown, but we suggest that DYX1C1 could provide a scaffold for macromolecular assembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3786.map.gz emd_3786.map.gz | 44 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3786-v30.xml emd-3786-v30.xml emd-3786.xml emd-3786.xml | 11.5 KB 11.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3786.png emd_3786.png | 133 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3786 http://ftp.pdbj.org/pub/emdb/structures/EMD-3786 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3786 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3786 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3786.map.gz / Format: CCP4 / Size: 47.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3786.map.gz / Format: CCP4 / Size: 47.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Subaverage after subtomogram classification of Chlamydomonas pf23 cilia, based on outer dynein arm structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.68 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
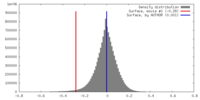
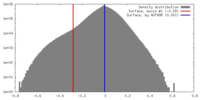
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Ciliary microtubule doublet with dynein, radial spokes and other ...
| Entire | Name: Ciliary microtubule doublet with dynein, radial spokes and other binding proteins |
|---|---|
| Components |
|
-Supramolecule #1: Ciliary microtubule doublet with dynein, radial spokes and other ...
| Supramolecule | Name: Ciliary microtubule doublet with dynein, radial spokes and other binding proteins type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Component - Name: HMDEK Details: 30 mM HEPES, 5 mM MgSO4, 1 mM DTT, 1 mM EGTA, and 50 mM potassium acetate, pH7.4 |
|---|---|
| Grid | Model: Plano lacey / Material: COPPER / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % / Chamber temperature: 298 K / Instrument: HOMEMADE PLUNGER / Details: one side blotting. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2200FS |
|---|---|
| Specialist optics | Energy filter - Name: JEOL in-column / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 25 eV |
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 2 / Average exposure time: 1.0 sec. / Average electron dose: 0.8 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 20000 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
- Image processing
Image processing
| Final reconstruction | Number classes used: 1 / Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 50.0 Å / Resolution method: OTHER / Software - Name: SPIDER Details: Visual estimation, compared with datasets in the past Number subtomograms used: 464 |
|---|---|
| Extraction | Number tomograms: 5 / Number images used: 660 |
| Final 3D classification | Number classes: 5 / Avg.num./class: 120 / Software - Name: Xmipp Details: This particular uploaded map is from 464 subtomograms. |
| Final angle assignment | Type: OTHER / Software - Name: SPIDER / Details: 3D cross correlation by Spider |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)