+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SpCas9-MMLV RT-pegRNA-target DNA complex (termination) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR-Cas / RNA BINDING PROTEIN-RNA-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell late endosome membrane / maintenance of CRISPR repeat elements / virion assembly / 3'-5' exonuclease activity / DNA endonuclease activity / host multivesicular body / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA-directed DNA polymerase activity ...host cell late endosome membrane / maintenance of CRISPR repeat elements / virion assembly / 3'-5' exonuclease activity / DNA endonuclease activity / host multivesicular body / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / defense response to virus / DNA recombination / structural constituent of virion / Hydrolases; Acting on ester bonds / aspartic-type endopeptidase activity / DNA-directed DNA polymerase activity / symbiont entry into host cell / host cell plasma membrane / proteolysis / DNA binding / RNA binding / zinc ion binding / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Streptococcus pyogenes (bacteria) / Streptococcus pyogenes (bacteria) /  Moloney murine leukemia virus / Moloney murine leukemia virus /  Streptococcus pyogenes serotype M1 (bacteria) Streptococcus pyogenes serotype M1 (bacteria) | |||||||||
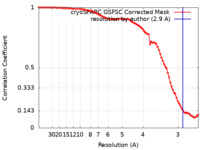
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Shuto Y / Nakagawa R / Hoki M / Omura SN / Hirano H / Itoh Y / Nureki O | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Structural basis for pegRNA-guided reverse transcription by a prime editor. Authors: Yutaro Shuto / Ryoya Nakagawa / Shiyou Zhu / Mizuki Hoki / Satoshi N Omura / Hisato Hirano / Yuzuru Itoh / Feng Zhang / Osamu Nureki /   Abstract: The prime editor system composed of Streptococcus pyogenes Cas9 nickase (nSpCas9) and engineered Moloney murine leukaemia virus reverse transcriptase (M-MLV RT) collaborates with a prime editing ...The prime editor system composed of Streptococcus pyogenes Cas9 nickase (nSpCas9) and engineered Moloney murine leukaemia virus reverse transcriptase (M-MLV RT) collaborates with a prime editing guide RNA (pegRNA) to facilitate a wide variety of precise genome edits in living cells. However, owing to a lack of structural information, the molecular mechanism of pegRNA-guided reverse transcription by the prime editor remains poorly understood. Here we present cryo-electron microscopy structures of the SpCas9-M-MLV RTΔRNaseH-pegRNA-target DNA complex in multiple states. The termination structure, along with our functional analysis, reveals that M-MLV RT extends reverse transcription beyond the expected site, resulting in scaffold-derived incorporations that cause undesired edits at the target loci. Furthermore, structural comparisons among the pre-initiation, initiation and elongation states show that M-MLV RT remains in a consistent position relative to SpCas9 during reverse transcription, whereas the pegRNA-synthesized DNA heteroduplex builds up along the surface of SpCas9. On the basis of our structural insights, we rationally engineered pegRNA variants and prime-editor variants in which M-MLV RT is fused within SpCas9. Collectively, our findings provide structural insights into the stepwise mechanism of prime editing, and will pave the way for the development of a versatile prime editing toolbox. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37858.map.gz emd_37858.map.gz | 17.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37858-v30.xml emd-37858-v30.xml emd-37858.xml emd-37858.xml | 20.3 KB 20.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37858_fsc.xml emd_37858_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_37858.png emd_37858.png | 120.9 KB | ||
| Masks |  emd_37858_msk_1.map emd_37858_msk_1.map | 19.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-37858.cif.gz emd-37858.cif.gz | 7.4 KB | ||
| Others |  emd_37858_half_map_1.map.gz emd_37858_half_map_1.map.gz emd_37858_half_map_2.map.gz emd_37858_half_map_2.map.gz | 18 MB 18 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37858 http://ftp.pdbj.org/pub/emdb/structures/EMD-37858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37858 | HTTPS FTP |
-Validation report
| Summary document |  emd_37858_validation.pdf.gz emd_37858_validation.pdf.gz | 820.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37858_full_validation.pdf.gz emd_37858_full_validation.pdf.gz | 819.7 KB | Display | |
| Data in XML |  emd_37858_validation.xml.gz emd_37858_validation.xml.gz | 14 KB | Display | |
| Data in CIF |  emd_37858_validation.cif.gz emd_37858_validation.cif.gz | 18.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37858 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37858 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37858 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37858 | HTTPS FTP |
-Related structure data
| Related structure data |  8wusMC  8wutC  8wuuC  8wuvC  8ygjC  39306  39307 M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37858.map.gz / Format: CCP4 / Size: 19.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37858.map.gz / Format: CCP4 / Size: 19.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2969 Å | ||||||||||||||||||||||||||||||||||||
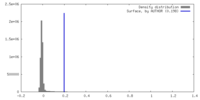
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_37858_msk_1.map emd_37858_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
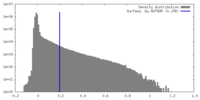
| Density Histograms |
-Half map: #2
| File | emd_37858_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
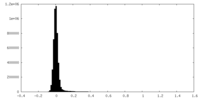
| Density Histograms |
-Half map: #1
| File | emd_37858_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
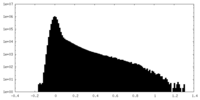
| Density Histograms |
- Sample components
Sample components
-Entire : SpCas9-MMLV RT-pegRNA-target DNA complex
| Entire | Name: SpCas9-MMLV RT-pegRNA-target DNA complex |
|---|---|
| Components |
|
-Supramolecule #1: SpCas9-MMLV RT-pegRNA-target DNA complex
| Supramolecule | Name: SpCas9-MMLV RT-pegRNA-target DNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
-Macromolecule #1: Gag-Pol polyprotein
| Macromolecule | Name: Gag-Pol polyprotein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Moloney murine leukemia virus Moloney murine leukemia virus |
| Molecular weight | Theoretical: 55.656242 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TLNIEDEYRL HETSKEPDVS LGSTWLSDFP QAWAETGGMG LAVRQAPLII PLKATSTPVS IKQYPMSQEA RLGIKPHIQR LLDQGILVP CQSPWNTPLL PVKKPGTNDY RPVQDLREVN KRVEDIHPTV PNPYNLLSGL PPSHQWYTVL DLKDAFFCLR L HPTSQPLF ...String: TLNIEDEYRL HETSKEPDVS LGSTWLSDFP QAWAETGGMG LAVRQAPLII PLKATSTPVS IKQYPMSQEA RLGIKPHIQR LLDQGILVP CQSPWNTPLL PVKKPGTNDY RPVQDLREVN KRVEDIHPTV PNPYNLLSGL PPSHQWYTVL DLKDAFFCLR L HPTSQPLF AFEWRDPEMG ISGQLTWTRL PQGFKNSPTL FNEALHRDLA DFRIQHPDLI LLQYVDDLLL AATSELDCQQ GT RALLQTL GNLGYRASAK KAQICQKQVK YLGYLLKEGQ RWLTEARKET VMGQPTPKTP RQLREFLGKA GFCRLFIPGF AEM AAPLYP LTKPGTLFNW GPDQQKAYQE IKQALLTAPA LGLPDLTKPF ELFVDEKQGY AKGVLTQKLG PWRRPVAYLS KKLD PVAAG WPPCLRMVAA IAVLTKDAGK LTMGQPLVIL APHAVEALVK QPPDRWLSNA RMTHYQALLL DTDRVQFGPV VALNP ATLL PLPEEGLQHN CL UniProtKB: Gag-Pol polyprotein |
-Macromolecule #2: CRISPR-associated endonuclease Cas9/Csn1
| Macromolecule | Name: CRISPR-associated endonuclease Cas9/Csn1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes serotype M1 (bacteria) Streptococcus pyogenes serotype M1 (bacteria) |
| Molecular weight | Theoretical: 158.457578 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DKKYSIGLAI GTNSVGWAVI TDEYKVPSKK FKVLGNTDRH SIKKNLIGAL LFDSGETAEA TRLKRTARRR YTRRKNRICY LQEIFSNEM AKVDDSFFHR LEESFLVEED KKHERHPIFG NIVDEVAYHE KYPTIYHLRK KLVDSTDKAD LRLIYLALAH M IKFRGHFL ...String: DKKYSIGLAI GTNSVGWAVI TDEYKVPSKK FKVLGNTDRH SIKKNLIGAL LFDSGETAEA TRLKRTARRR YTRRKNRICY LQEIFSNEM AKVDDSFFHR LEESFLVEED KKHERHPIFG NIVDEVAYHE KYPTIYHLRK KLVDSTDKAD LRLIYLALAH M IKFRGHFL IEGDLNPDNS DVDKLFIQLV QTYNQLFEEN PINASGVDAK AILSARLSKS RRLENLIAQL PGEKKNGLFG NL IALSLGL TPNFKSNFDL AEDAKLQLSK DTYDDDLDNL LAQIGDQYAD LFLAAKNLSD AILLSDILRV NTEITKAPLS ASM IKRYDE HHQDLTLLKA LVRQQLPEKY KEIFFDQSKN GYAGYIDGGA SQEEFYKFIK PILEKMDGTE ELLVKLNRED LLRK QRTFD NGSIPHQIHL GELHAILRRQ EDFYPFLKDN REKIEKILTF RIPYYVGPLA RGNSRFAWMT RKSEETITPW NFEEV VDKG ASAQSFIERM TNFDKNLPNE KVLPKHSLLY EYFTVYNELT KVKYVTEGMR KPAFLSGEQK KAIVDLLFKT NRKVTV KQL KEDYFKKIEC FDSVEISGVE DRFNASLGTY HDLLKIIKDK DFLDNEENED ILEDIVLTLT LFEDREMIEE RLKTYAH LF DDKVMKQLKR RRYTGWGRLS RKLINGIRDK QSGKTILDFL KSDGFANRNF MQLIHDDSLT FKEDIQKAQV SGQGDSLH E HIANLAGSPA IKKGILQTVK VVDELVKVMG RHKPENIVIE MARENQTTQK GQKNSRERMK RIEEGIKELG SQILKEHPV ENTQLQNEKL YLYYLQNGRD MYVDQELDIN RLSDYDVDAI VPQSFLKDDS IDNKVLTRSD KNRGKSDNVP SEEVVKKMKN YWRQLLNAK LITQRKFDNL TKAERGGLSE LDKAGFIKRQ LVETRQITKH VAQILDSRMN TKYDENDKLI REVKVITLKS K LVSDFRKD FQFYKVREIN NYHHAHDAYL NAVVGTALIK KYPKLESEFV YGDYKVYDVR KMIAKSEQEI GKATAKYFFY SN IMNFFKT EITLANGEIR KRPLIETNGE TGEIVWDKGR DFATVRKVLS MPQVNIVKKT EVQTGGFSKE SILPKRNSDK LIA RKKDWD PKKYGGFDSP TVAYSVLVVA KVEKGKSKKL KSVKELLGIT IMERSSFEKN PIDFLEAKGY KEVKKDLIIK LPKY SLFEL ENGRKRMLAS AGELQKGNEL ALPSKYVNFL YLASHYEKLK GSPEDNEQKQ LFVEQHKHYL DEIIEQISEF SKRVI LADA NLDKVLSAYN KHRDKPIREQ AENIIHLFTL TNLGAPAAFK YFDTTIDRKR YTSTKEVLDA TLIHQSITGL YETRID LSQ LGGD UniProtKB: CRISPR-associated endonuclease Cas9/Csn1 |
-Macromolecule #3: DNA (51-MER)
| Macromolecule | Name: DNA (51-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
| Molecular weight | Theoretical: 15.636033 KDa |
| Sequence | String: (DC)(DT)(DA)(DG)(DT)(DA)(DC)(DT)(DC)(DT) (DG)(DC)(DC)(DA)(DT)(DC)(DA)(DG)(DA)(DG) (DC)(DA)(DA)(DG)(DC)(DA)(DC)(DT)(DA) (DC)(DG)(DG)(DC)(DC)(DG)(DA)(DT)(DT)(DG) (DC) (DT)(DC)(DT)(DA)(DA)(DG)(DT)(DG) (DA)(DT)(DC) |
-Macromolecule #4: DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3')
| Macromolecule | Name: DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3') type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
| Molecular weight | Theoretical: 5.291446 KDa |
| Sequence | String: (DT)(DG)(DA)(DT)(DG)(DG)(DC)(DA)(DG)(DA) (DG)(DT)(DA)(DC)(DT)(DA)(DG) |
-Macromolecule #6: DNA (40-MER)
| Macromolecule | Name: DNA (40-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
| Molecular weight | Theoretical: 12.315929 KDa |
| Sequence | String: (DG)(DA)(DT)(DC)(DA)(DC)(DT)(DT)(DA)(DG) (DA)(DG)(DC)(DA)(DA)(DT)(DC)(DG)(DG)(DC) (DC)(DC)(DA)(DG)(DA)(DC)(DT)(DG)(DA) (DG)(DC)(DA)(DC)(DG)(DC)(DT)(DG)(DT)(DG) (2DA) |
-Macromolecule #5: RNA (115-MER)
| Macromolecule | Name: RNA (115-MER) / type: rna / ID: 5 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
| Molecular weight | Theoretical: 37.065949 KDa |
| Sequence | String: GGCCGUAGUG CUUGCUCUGA GUUUUAGAGC UAGAAAUAGC AAGUUAAAAU AAGGCUAGUC CGUUAUCAAC UUGAAAAAGU GGCACCGAG UCGGUGCUCA CAGCGUGCUC AGUCUG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 297 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)