+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of human TMEM63A | |||||||||
 Map data Map data | sharpen map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mechanically activated (MA) ion channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsurfactant secretion / osmolarity-sensing monoatomic cation channel activity / mechanosensitive monoatomic ion channel activity / calcium-activated cation channel activity / tertiary granule membrane / specific granule membrane / centriolar satellite / early endosome membrane / nucleic acid binding / lysosomal membrane ...surfactant secretion / osmolarity-sensing monoatomic cation channel activity / mechanosensitive monoatomic ion channel activity / calcium-activated cation channel activity / tertiary granule membrane / specific granule membrane / centriolar satellite / early endosome membrane / nucleic acid binding / lysosomal membrane / intracellular membrane-bounded organelle / Neutrophil degranulation / extracellular exosome / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Yang D | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Proteins / Year: 2024 Journal: Proteins / Year: 2024Title: A monomeric structure of human TMEM63A protein. Authors: Xuening Wu / Tiantian Shang / Xinyi Lü / Deyi Luo / Dongxue Yang /  Abstract: OSCA/TMEM63 is a newly identified family of mechanically activated (MA) ion channels in plants and animals, respectively, which convert physical forces into electrical signals or trigger ...OSCA/TMEM63 is a newly identified family of mechanically activated (MA) ion channels in plants and animals, respectively, which convert physical forces into electrical signals or trigger intracellular cascades and are essential for eukaryotic physiology. OSCAs and related TMEM16s and transmembrane channel-like (TMC) proteins form homodimers with two pores. However, the molecular architecture of the mammalian TMEM63 proteins remains unclear. Here we elucidate the structure of human TMEM63A in the presence of calcium by single particle cryo-EM, revealing a distinct monomeric architecture containing eleven transmembrane helices. It has structural similarity to the single subunit of the Arabidopsis thaliana OSCA proteins. We locate the ion permeation pathway within the monomeric configuration and observe a nonprotein density resembling lipid. These results lay a foundation for understanding the structural organization of OSCA/TMEM63A family proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37852.map.gz emd_37852.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37852-v30.xml emd-37852-v30.xml emd-37852.xml emd-37852.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37852.png emd_37852.png | 100.4 KB | ||
| Filedesc metadata |  emd-37852.cif.gz emd-37852.cif.gz | 6.2 KB | ||
| Others |  emd_37852_additional_1.map.gz emd_37852_additional_1.map.gz emd_37852_half_map_1.map.gz emd_37852_half_map_1.map.gz emd_37852_half_map_2.map.gz emd_37852_half_map_2.map.gz | 31.8 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37852 http://ftp.pdbj.org/pub/emdb/structures/EMD-37852 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37852 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37852 | HTTPS FTP |
-Validation report
| Summary document |  emd_37852_validation.pdf.gz emd_37852_validation.pdf.gz | 798.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37852_full_validation.pdf.gz emd_37852_full_validation.pdf.gz | 798.2 KB | Display | |
| Data in XML |  emd_37852_validation.xml.gz emd_37852_validation.xml.gz | 12.3 KB | Display | |
| Data in CIF |  emd_37852_validation.cif.gz emd_37852_validation.cif.gz | 14.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37852 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37852 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37852 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37852 | HTTPS FTP |
-Related structure data
| Related structure data |  8wuaMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_37852.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37852.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpen map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0979 Å | ||||||||||||||||||||||||||||||||||||
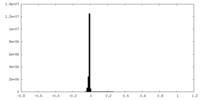
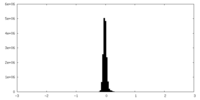
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: unsharpen map
| File | emd_37852_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpen map | ||||||||||||
| Projections & Slices |
| ||||||||||||
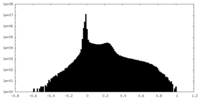
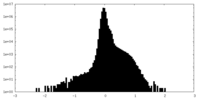
| Density Histograms |
-Half map: half map A
| File | emd_37852_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
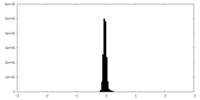
| Density Histograms |
-Half map: half map B
| File | emd_37852_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
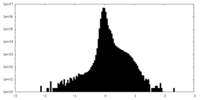
| Density Histograms |
- Sample components
Sample components
-Entire : human TMEM63A
| Entire | Name: human TMEM63A |
|---|---|
| Components |
|
-Supramolecule #1: human TMEM63A
| Supramolecule | Name: human TMEM63A / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: CSC1-like protein 1
| Macromolecule | Name: CSC1-like protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 82.331828 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: FLELWQSKAV SIREQLGLGD RPNDSYCYNS AKNSTVLQGV TFGGIPTVLL IDVSCFLFLI LVFSIIRRRF WDYGRIALVS EADSESRFQ RLSSTSSSGQ QDFENELGCC PWLTAIFRLH DDQILEWCGE DAIHYLSFQR HIIFLLVVVS FLSLCVILPV N LSGDLLDK ...String: FLELWQSKAV SIREQLGLGD RPNDSYCYNS AKNSTVLQGV TFGGIPTVLL IDVSCFLFLI LVFSIIRRRF WDYGRIALVS EADSESRFQ RLSSTSSSGQ QDFENELGCC PWLTAIFRLH DDQILEWCGE DAIHYLSFQR HIIFLLVVVS FLSLCVILPV N LSGDLLDK DPYSFGRTTI ANLQTDNDLL WLHTIFAVIY LFLTVGFMRH HTQSIKYKEE NLVRRTLFIT GLPRDARKET VE SHFRDAY PTCEVVDVQL CYNVAKLIYL CKEKKKTEKS LTYYTNLQVK TGQRTLINPK PCGQFCCCEV LGCEWEDAIS YYT RMKDRL LERITEEERH VQDQPLGMAF VTFQEKSMAT YILKDFNACK CQSLQCKGEP QPSSHSRELY TSKWTVTFAA DPED ICWKN LSIQGLRWWL QWLGINFTLF LGLFFLTTPS IILSTMDKFN VTKPIHALNN PIISQFFPTL LLWSFSALLP SIVYY STLL ESHWTKSGEN QIMMTKVYIF LIFMVLILPS LGLTSLDFFF RWLFDKTSSE ASIRLECVFL PDQGAFFVNY VIASAF IGN GMELLRLPGL ILYTFRMIMA KTAADRRNVK QNQAFQYEFG AMYAWMLCVF TVIVAYSITC PIIAPFGLIY ILLKHMV DR HNLYFVYLPA KLEKGIHFAA VNQALAAPIL CLFWLYFFSF LRLGMKAPAT LFTFLVLLLT ILVCLAHTCF GC UniProtKB: CSC1-like protein 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.56 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)