[English] 日本語
 Yorodumi
Yorodumi- EMDB-37795: Cry-EM structure of cannabinoid receptor-beta-arrestin-1 complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cry-EM structure of cannabinoid receptor-beta-arrestin-1 complex | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | complex / MEMBRANE PROTEIN / GPCR | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcannabinoid signaling pathway / renal water retention / Defective AVP does not bind AVPR2 and causes neurohypophyseal diabetes insipidus (NDI) / retrograde trans-synaptic signaling by endocannabinoid / Vasopressin-like receptors / regulation of systemic arterial blood pressure by vasopressin / vasopressin receptor activity / cannabinoid receptor activity / angiotensin receptor binding / TGFBR3 regulates TGF-beta signaling ...cannabinoid signaling pathway / renal water retention / Defective AVP does not bind AVPR2 and causes neurohypophyseal diabetes insipidus (NDI) / retrograde trans-synaptic signaling by endocannabinoid / Vasopressin-like receptors / regulation of systemic arterial blood pressure by vasopressin / vasopressin receptor activity / cannabinoid receptor activity / angiotensin receptor binding / TGFBR3 regulates TGF-beta signaling / hemostasis / Activation of SMO / regulation of presynaptic cytosolic calcium ion concentration / telencephalon development / negative regulation of interleukin-8 production / Class A/1 (Rhodopsin-like receptors) / arrestin family protein binding / axonal fasciculation / G protein-coupled receptor internalization / Lysosome Vesicle Biogenesis / : / stress fiber assembly / positive regulation of cardiac muscle hypertrophy / Golgi Associated Vesicle Biogenesis / positive regulation of Rho protein signal transduction / pseudopodium / positive regulation of systemic arterial blood pressure / negative regulation of interleukin-6 production / positive regulation of intracellular signal transduction / positive regulation of receptor internalization / positive regulation of vasoconstriction / negative regulation of Notch signaling pathway / endocytic vesicle / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / activation of adenylate cyclase activity / cellular response to hormone stimulus / insulin-like growth factor receptor binding / response to cytokine / clathrin-coated pit / negative regulation of protein ubiquitination / enzyme inhibitor activity / GTPase activator activity / Activated NOTCH1 Transmits Signal to the Nucleus / cytoplasmic vesicle membrane / clathrin-coated endocytic vesicle membrane / Signaling by high-kinase activity BRAF mutants / electron transport chain / MAP2K and MAPK activation / G protein-coupled receptor binding / G protein-coupled receptor activity / GABA-ergic synapse / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / positive regulation of protein phosphorylation / Signaling by RAF1 mutants / Signaling by moderate kinase activity BRAF mutants / Paradoxical activation of RAF signaling by kinase inactive BRAF / Signaling downstream of RAS mutants / endocytic vesicle membrane / Signaling by BRAF and RAF1 fusions / Vasopressin regulates renal water homeostasis via Aquaporins / Cargo recognition for clathrin-mediated endocytosis / actin cytoskeleton / glucose homeostasis / protein transport / Clathrin-mediated endocytosis / Thrombin signalling through proteinase activated receptors (PARs) / growth cone / presynaptic membrane / cytoplasmic vesicle / G alpha (i) signalling events / G alpha (s) signalling events / ubiquitin-dependent protein catabolic process / molecular adaptor activity / proteasome-mediated ubiquitin-dependent protein catabolic process / mitochondrial outer membrane / transcription coactivator activity / electron transfer activity / periplasmic space / positive regulation of ERK1 and ERK2 cascade / endosome / Ub-specific processing proteases / nuclear body / protein ubiquitination / iron ion binding / membrane raft / G protein-coupled receptor signaling pathway / Golgi membrane / negative regulation of cell population proliferation / lysosomal membrane / heme binding / positive regulation of cell population proliferation / ubiquitin protein ligase binding / positive regulation of gene expression / regulation of transcription by RNA polymerase II / chromatin / perinuclear region of cytoplasm / glutamatergic synapse / endoplasmic reticulum / Golgi apparatus Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) / Phage display vector pTDisp (others) Homo sapiens (human) / Phage display vector pTDisp (others) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | ||||||||||||||||||
 Authors Authors | Wang YX / Wang T / Wu LJ / Hua T / Liu ZJ | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Protein Cell / Year: 2024 Journal: Protein Cell / Year: 2024Title: Cryo-EM structure of cannabinoid receptor CB1-β-arrestin complex. Authors: Yuxia Wang / Lijie Wu / Tian Wang / Junlin Liu / Fei Li / Longquan Jiang / Zhongbo Fan / Yanan Yu / Na Chen / Qianqian Sun / Qiwen Tan / Tian Hua / Zhi-Jie Liu /  | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37795.map.gz emd_37795.map.gz | 57 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37795-v30.xml emd-37795-v30.xml emd-37795.xml emd-37795.xml | 34.6 KB 34.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37795.png emd_37795.png | 34.5 KB | ||
| Filedesc metadata |  emd-37795.cif.gz emd-37795.cif.gz | 8.8 KB | ||
| Others |  emd_37795_half_map_1.map.gz emd_37795_half_map_1.map.gz emd_37795_half_map_2.map.gz emd_37795_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37795 http://ftp.pdbj.org/pub/emdb/structures/EMD-37795 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37795 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37795 | HTTPS FTP |
-Related structure data
| Related structure data |  8wrzMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37795.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37795.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
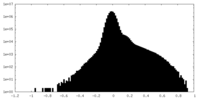
-Half map: #2
| File | emd_37795_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
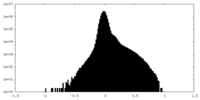
-Half map: #1
| File | emd_37795_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cry-EM structure of cannabinoid receptor-arrestin 1 complex
| Entire | Name: Cry-EM structure of cannabinoid receptor-arrestin 1 complex |
|---|---|
| Components |
|
-Supramolecule #1: Cry-EM structure of cannabinoid receptor-arrestin 1 complex
| Supramolecule | Name: Cry-EM structure of cannabinoid receptor-arrestin 1 complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Beta-arrestin-1
| Macromolecule | Name: Beta-arrestin-1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 44.271508 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGDKGTRVFK KASPNGKLTV YLGKRDFVDH IDLVDPVDGV VLVDPEYLKE RRVYVTLTCA FRYGREDLDV LGLTFRKDLF VANVQSFPP APEDKKPLTR LQERLIKKLG EHAYPFTFEI PPNLPCSVTL QPGPEDTGKA CGVDYEVKAF CAENLEEKIH K RNSVRLVI ...String: MGDKGTRVFK KASPNGKLTV YLGKRDFVDH IDLVDPVDGV VLVDPEYLKE RRVYVTLTCA FRYGREDLDV LGLTFRKDLF VANVQSFPP APEDKKPLTR LQERLIKKLG EHAYPFTFEI PPNLPCSVTL QPGPEDTGKA CGVDYEVKAF CAENLEEKIH K RNSVRLVI RKVQYAPERP GPQPTAETTR QFLMSDKPLH LEASLDKEIY YHGEPISVNV HVTNNTNKTV KKIKISVRQY AD ICLFNTA QYKCPVAMEE ADDTVAPSST FCKVYTLTPF LANNREKRGL ALDGKLKHED TNLASSTLLR EGANREILGI IVS YKVKVK LVVSRGGLLG DLASSDVAVE LPFTLMHPKP KEEPPHREVP ENETPVDTNL IELDTNDDDI VFEDFAR UniProtKB: Beta-arrestin-1 |
-Macromolecule #2: scfv30 light chain, scfv30 heavy chain
| Macromolecule | Name: scfv30 light chain, scfv30 heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: Phage display vector pTDisp (others) |
| Molecular weight | Theoretical: 28.693922 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKKNIAFLLA SMFVFSIATN AYASDIQMTQ SPSSLSASVG DRVTITCRAS QSVSSAVAWY QQKPGKAPKL LIYSASSLYS GVPSRFSGS RSGTDFTLTI SSLQPEDFAT YYCQQYKYVP VTFGQGTKVE IKGTTAASGS SGGSSSGAEV QLVESGGGLV Q PGGSLRLS ...String: MKKNIAFLLA SMFVFSIATN AYASDIQMTQ SPSSLSASVG DRVTITCRAS QSVSSAVAWY QQKPGKAPKL LIYSASSLYS GVPSRFSGS RSGTDFTLTI SSLQPEDFAT YYCQQYKYVP VTFGQGTKVE IKGTTAASGS SGGSSSGAEV QLVESGGGLV Q PGGSLRLS CAASGFNVYS SSIHWVRQAP GKGLEWVASI SSYYGYTYYA DSVKGRFTIS ADTSKNTAYL QMNSLRAEDT AV YYCARSR QFWYSGLDYW GQGTLVTVSS A |
-Macromolecule #3: Soluble cytochrome b562,Cannabinoid receptor 1
| Macromolecule | Name: Soluble cytochrome b562,Cannabinoid receptor 1 / type: protein_or_peptide / ID: 3 Details: The chimera of Cytochrome b-562, linker, and Cannabinoid receptor 1 Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 53.577543 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ADLEDNWETL NDNLKVIEKA DNAAQVKDAL TKMRAAALDA QKATPPKLED KSPDSPEMKD FRHGFDILVG QIDDALKLAN EGKVKEAQA AAEQLKTTRN AYIQKYLLEV LFQGPVPADQ VNITEFYNKS LSSFKENEEN IQCGENFMDI ECFMVLNPSQ Q LAIAVLSL ...String: ADLEDNWETL NDNLKVIEKA DNAAQVKDAL TKMRAAALDA QKATPPKLED KSPDSPEMKD FRHGFDILVG QIDDALKLAN EGKVKEAQA AAEQLKTTRN AYIQKYLLEV LFQGPVPADQ VNITEFYNKS LSSFKENEEN IQCGENFMDI ECFMVLNPSQ Q LAIAVLSL TLGTFTVLEN LLVLCVILHS RSLRCRPSYH FIGSLAVADL LGSVIFVYSF IDFHVFHRKD SRNVFLFKLG GV TASFTAS VGSLFLIAID RYISIHRPLA YKRIVTRPKA VVAFCLMWTI AIVIAVLPLL GWNCEKLQSV CSDIFPHIDK TYL MFWIGV VSVLLLFIVY AYMYILWKAH SHAVRMIQRG TQKSIIIHTS EDGKVQVTRP DQARMDIELA KTLVLILVVL IICW GPLLA IMVYDVFGKM NKLIKTVFAF CSMLCLLNST VNPIIYALRS KDLRHAFRSM FPSCEGTAQP LDNSMGDSDC L UniProtKB: Soluble cytochrome b562, Cannabinoid receptor 1 |
-Macromolecule #4: Vasopressin V2 receptor
| Macromolecule | Name: Vasopressin V2 receptor / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.920651 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ARGRTPPSLG PQDESC(TPO)(TPO)A(SEP) (SEP)(SEP)LAK UniProtKB: Vasopressin V2 receptor |
-Macromolecule #5: (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanat...
| Macromolecule | Name: (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol type: ligand / ID: 5 / Number of copies: 1 / Formula: 8D0 |
|---|---|
| Molecular weight | Theoretical: 445.658 Da |
| Chemical component information | 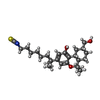 ChemComp-8D0: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 25847 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8wrz: |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)