+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3772 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | hINO80 core complex | |||||||||
 Map data Map data | Low resolution INO80 core EM map. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
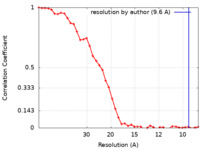
| Method | single particle reconstruction / cryo EM / Resolution: 9.6 Å | |||||||||
 Authors Authors | Aramayo RJ / Willhoft O / Ayala R / Bythell-Douglas R / Wigley D / Zhang X | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2018 Journal: Nat Struct Mol Biol / Year: 2018Title: Cryo-EM structures of the human INO80 chromatin-remodeling complex. Authors: Ricardo J Aramayo / Oliver Willhoft / Rafael Ayala / Rohan Bythell-Douglas / Dale B Wigley / Xiaodong Zhang /  Abstract: Access to chromatin for processes such as transcription and DNA repair requires the sliding of nucleosomes along DNA. This process is aided by chromatin-remodeling complexes, such as the multisubunit ...Access to chromatin for processes such as transcription and DNA repair requires the sliding of nucleosomes along DNA. This process is aided by chromatin-remodeling complexes, such as the multisubunit INO80 chromatin-remodeling complex. Here we present cryo-EM structures of the active core complex of human INO80 at 9.6 Å, with portions at 4.1-Å resolution, and reconstructions of combinations of subunits. Together, these structures reveal the architecture of the INO80 complex, including Ino80 and actin-related proteins, which is assembled around a single RUVBL1 (Tip49a) and RUVBL2 (Tip49b) AAA+ heterohexamer. An unusual spoked-wheel structural domain of the Ino80 subunit is engulfed by this heterohexamer; both, in combination, form the core of the complex. We also identify a cleft in RUVBL1 and RUVBL2, which forms a major interaction site for partner proteins and probably communicates these interactions to its nucleotide-binding sites. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3772.map.gz emd_3772.map.gz | 28.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3772-v30.xml emd-3772-v30.xml emd-3772.xml emd-3772.xml | 11.4 KB 11.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3772_fsc.xml emd_3772_fsc.xml | 4.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_3772.png emd_3772.png | 43.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3772 http://ftp.pdbj.org/pub/emdb/structures/EMD-3772 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3772 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3772 | HTTPS FTP |
-Validation report
| Summary document |  emd_3772_validation.pdf.gz emd_3772_validation.pdf.gz | 260.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3772_full_validation.pdf.gz emd_3772_full_validation.pdf.gz | 259.3 KB | Display | |
| Data in XML |  emd_3772_validation.xml.gz emd_3772_validation.xml.gz | 8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3772 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3772 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3772 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3772 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3772.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3772.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Low resolution INO80 core EM map. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.26 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human INO80 core complex
| Entire | Name: Human INO80 core complex |
|---|---|
| Components |
|
-Supramolecule #1: Human INO80 core complex
| Supramolecule | Name: Human INO80 core complex / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 800 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation #1
Sample preparation #1
| Preparation ID | 1 |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Film type ID: 1 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Sample preparation #2
Sample preparation #2
| Preparation ID | 2 |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Film type ID: 1 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TITAN KRIOS |
| Image recording | Image recording ID: 1 / Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Electron microscopy #1~
Electron microscopy #1~
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TITAN KRIOS |
| Image recording | Image recording ID: 2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)