+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ADP-bound purinergic receptor 1 in complex with miniGs/q | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationG protein-coupled ATP receptor activity / cellular response to purine-containing compound / relaxation of muscle / G protein-coupled ADP receptor activity / A1 adenosine receptor binding / G protein-coupled purinergic nucleotide receptor signaling pathway / positive regulation of inositol trisphosphate biosynthetic process / P2Y receptors / positive regulation of penile erection / G protein-coupled purinergic nucleotide receptor activity ...G protein-coupled ATP receptor activity / cellular response to purine-containing compound / relaxation of muscle / G protein-coupled ADP receptor activity / A1 adenosine receptor binding / G protein-coupled purinergic nucleotide receptor signaling pathway / positive regulation of inositol trisphosphate biosynthetic process / P2Y receptors / positive regulation of penile erection / G protein-coupled purinergic nucleotide receptor activity / negative regulation of norepinephrine secretion / positive regulation of monoatomic ion transport / glial cell migration / signaling receptor regulator activity / regulation of presynaptic cytosolic calcium ion concentration / G protein-coupled adenosine receptor signaling pathway / response to growth factor / positive regulation of hormone secretion / signal transduction involved in regulation of gene expression / eating behavior / regulation of synaptic vesicle exocytosis / cellular response to ATP / response to mechanical stimulus / monoatomic ion transport / presynaptic active zone membrane / blood vessel diameter maintenance / protein localization to plasma membrane / establishment of localization in cell / electron transport chain / ADP binding / platelet activation / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / regulation of cell shape / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / signaling receptor activity / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / positive regulation of cytosolic calcium ion concentration / cell body / GTPase binding / Ca2+ pathway / fibroblast proliferation / scaffold protein binding / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / basolateral plasma membrane / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / postsynaptic membrane / electron transfer activity / periplasmic space / cell surface receptor signaling pathway / Extra-nuclear estrogen signaling / positive regulation of ERK1 and ERK2 cascade / cell population proliferation / apical plasma membrane / postsynaptic density / cilium / iron ion binding / G protein-coupled receptor signaling pathway / protein heterodimerization activity / lysosomal membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Gu QC / Tang WQ | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: ADP-bound purinergic receptor 1 in complex with miniGs/q Authors: Gu QC / Tang WQ #1:  Journal: To Be Published Journal: To Be PublishedTitle: Real-space refinement in PHENIX for cryo-EM and crystallography Authors: Gu QC / Tang WQ | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37591.map.gz emd_37591.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37591-v30.xml emd-37591-v30.xml emd-37591.xml emd-37591.xml | 26.6 KB 26.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37591_fsc.xml emd_37591_fsc.xml | 7.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_37591.png emd_37591.png | 31.4 KB | ||
| Masks |  emd_37591_msk_1.map emd_37591_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-37591.cif.gz emd-37591.cif.gz | 7.9 KB | ||
| Others |  emd_37591_half_map_1.map.gz emd_37591_half_map_1.map.gz emd_37591_half_map_2.map.gz emd_37591_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37591 http://ftp.pdbj.org/pub/emdb/structures/EMD-37591 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37591 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37591 | HTTPS FTP |
-Related structure data
| Related structure data |  8wjxMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37591.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37591.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_37591_msk_1.map emd_37591_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
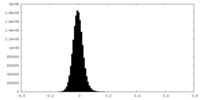
| Density Histograms |
-Half map: #2
| File | emd_37591_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
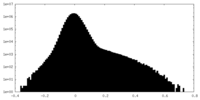
| Density Histograms |
-Half map: #1
| File | emd_37591_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ADP-bound purinergic receptor 1 in complex with miniGs/q
| Entire | Name: ADP-bound purinergic receptor 1 in complex with miniGs/q |
|---|---|
| Components |
|
-Supramolecule #1: ADP-bound purinergic receptor 1 in complex with miniGs/q
| Supramolecule | Name: ADP-bound purinergic receptor 1 in complex with miniGs/q type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: GNAS complex locus
| Macromolecule | Name: GNAS complex locus / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 30.840807 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGMGHHHHHH ENLYFQGNSK TEDQRNEEKA QREANKKIEK QLQKDKQVYR ATHRLLLLGA DNSGKSTIVK QMRILHGGSG GSGGTSGIF ETKFQVDKVN FHMFDVGGQR DERRKWIQCF NDVTAIIFVV DSSDYNRLQE ALNLFKSIWN NRWLRTISVI L FLNKQDLL ...String: MGMGHHHHHH ENLYFQGNSK TEDQRNEEKA QREANKKIEK QLQKDKQVYR ATHRLLLLGA DNSGKSTIVK QMRILHGGSG GSGGTSGIF ETKFQVDKVN FHMFDVGGQR DERRKWIQCF NDVTAIIFVV DSSDYNRLQE ALNLFKSIWN NRWLRTISVI L FLNKQDLL AEKVLAGKSK IEDYFPEFAR YTTPEDATPE PGEDPRVTRA KYFIRDEFLR ISTASGDGRH YCYPHFTCAV DT ENARRIF NDCKDIILQM NLREYNLV |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.055867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI ...String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CRQTFTGHES DI NAICFFP NGNAFATGSD DATCRLFDLR ADQELMTYSH DNIICGITSV SFSKSGRLLL AGYDDFNCNV WDALKADRAG VLA GHDNRV SCLGVTDDGM AVATGSWDSF LKIWNGSSGG GGSGGGGSSG VSGWRLFKKI S UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Nanobody 35
| Macromolecule | Name: Nanobody 35 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 17.057271 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKYLLPTAAA GLLLLAAQPA MAMQVQLQES GGGLVQPGGS LRLSCAASGF TFSNYKMNWV RQAPGKGLEW VSDISQSGAS ISYTGSVKG RFTISRDNAK NTLYLQMNSL KPEDTAVYYC ARCPAPFTRD CFDVTSTTYA YRGQGTQVTV SSHHHHHH |
-Macromolecule #5: Soluble cytochrome b562,P2Y purinoceptor 1
| Macromolecule | Name: Soluble cytochrome b562,P2Y purinoceptor 1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 76.586242 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDAGRAHHH HHHHHHHENL YFQSGAPADL EDNWETLNDN LKVIEKADNA AQVKDALTKM RAAALDAQK ATPPKLEDKS PDSPEMKDFR HGFDILVGQI DDALKLANEG KVKEAQAAAE QLKTTRNAYI QKYLEFLEVL F QGPTEVLW ...String: MKTIIALSYI FCLVFADYKD DDDAGRAHHH HHHHHHHENL YFQSGAPADL EDNWETLNDN LKVIEKADNA AQVKDALTKM RAAALDAQK ATPPKLEDKS PDSPEMKDFR HGFDILVGQI DDALKLANEG KVKEAQAAAE QLKTTRNAYI QKYLEFLEVL F QGPTEVLW PAVPNGTDAA FLAGPGSSWG NSTVASTAAV SSSFKCALTK TGFQFYYLPA VYILVFIIGF LGNSVAIWMF VF HMKPWSG ISVYMFNLAL ADFLYVLTLP ALIFYYFNKT DWIFGDAMCK LQRFIFHVNL YGSILFLTCI SAHRYSGVVY PLK SLGRLK KKNAICISVL VWLIVVVAIS PILFYSGTGV RKNKTITCYD TTSDEYLRSY FIYSMCTTVA MFCVPLVLIL GCYG LIVRA LIYKDLDNSP LRRKSIYLVI IVLTVFAVSY IPFHVMKTMN LRARLDFQTP AMCAFNDRVY ATYQVTRGLA SLNSC VDPI LYFLAGDTFR RRLSRATRKA SRRSEANLQS KSEDMTLNVF TLEDFVGDWE QTAAYNLDQV LEQGGVSSLL QNLAVS VTP IQRIVRSGEN ALKIDIHVII PYEGLSADQM AQIEEVFKVV YPVDDHHFKV ILPYGTLVID GVTPNMLNYF GRPYEGI AV FDGKKITVTG TLWNGNKIID ERLITPDGSM LFRVTINS UniProtKB: Soluble cytochrome b562, P2Y purinoceptor 1 |
-Macromolecule #6: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #7: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 7 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)