+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | SARS-CoV-2 Omicron XBB.1.5 RBD complexed with human ACE2 and S304 | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | RBD / VIRAL PROTEIN / VIRUS / VIRUS-IMMUNE SYSTEM complex | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報positive regulation of amino acid transport / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / 加水分解酵素; プロテアーゼ; ペプチド結合加水分解酵素; 金属プロテアーゼ / angiotensin-mediated drinking behavior / regulation of systemic arterial blood pressure by renin-angiotensin / tryptophan transport / positive regulation of gap junction assembly / regulation of cardiac conduction / regulation of vasoconstriction ...positive regulation of amino acid transport / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / 加水分解酵素; プロテアーゼ; ペプチド結合加水分解酵素; 金属プロテアーゼ / angiotensin-mediated drinking behavior / regulation of systemic arterial blood pressure by renin-angiotensin / tryptophan transport / positive regulation of gap junction assembly / regulation of cardiac conduction / regulation of vasoconstriction / peptidyl-dipeptidase activity / maternal process involved in female pregnancy / angiotensin maturation / Metabolism of Angiotensinogen to Angiotensins / negative regulation of signaling receptor activity / carboxypeptidase activity / Attachment and Entry / positive regulation of cardiac muscle contraction / viral life cycle / regulation of cytokine production / blood vessel diameter maintenance / negative regulation of smooth muscle cell proliferation / brush border membrane / regulation of transmembrane transporter activity / cilium / negative regulation of ERK1 and ERK2 cascade / metallopeptidase activity / positive regulation of reactive oxygen species metabolic process / endocytic vesicle membrane / virus receptor activity / regulation of cell population proliferation / regulation of inflammatory response / endopeptidase activity / Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Potential therapeutics for SARS / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / membrane fusion / receptor-mediated endocytosis of virus by host cell / Attachment and Entry / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / receptor ligand activity / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / symbiont entry into host cell / membrane raft / apical plasma membrane / endoplasmic reticulum lumen / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / cell surface / extracellular space / extracellular exosome / zinc ion binding / extracellular region / identical protein binding / membrane / plasma membrane 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) / Homo sapiens (ヒト) /  | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.91 Å | |||||||||
 データ登録者 データ登録者 | Li W / Xie Y | |||||||||
| 資金援助 |  中国, 1件 中国, 1件
| |||||||||
 引用 引用 |  ジャーナル: EMBO J / 年: 2024 ジャーナル: EMBO J / 年: 2024タイトル: Key mechanistic features of the trade-off between antibody escape and host cell binding in the SARS-CoV-2 Omicron variant spike proteins. 著者: Weiwei Li / Zepeng Xu / Tianhui Niu / Yufeng Xie / Zhennan Zhao / Dedong Li / Qingwen He / Wenqiao Sun / Kaiyuan Shi / Wenjing Guo / Zhen Chang / Kefang Liu / Zheng Fan / Jianxun Qi / George F Gao /  要旨: Since SARS-CoV-2 Omicron variant emerged, it is constantly evolving into multiple sub-variants, including BF.7, BQ.1, BQ.1.1, XBB, XBB.1.5 and the recently emerged BA.2.86 and JN.1. Receptor binding ...Since SARS-CoV-2 Omicron variant emerged, it is constantly evolving into multiple sub-variants, including BF.7, BQ.1, BQ.1.1, XBB, XBB.1.5 and the recently emerged BA.2.86 and JN.1. Receptor binding and immune evasion are recognized as two major drivers for evolution of the receptor binding domain (RBD) of the SARS-CoV-2 spike (S) protein. However, the underlying mechanism of interplay between two factors remains incompletely understood. Herein, we determined the structures of human ACE2 complexed with BF.7, BQ.1, BQ.1.1, XBB and XBB.1.5 RBDs. Based on the ACE2/RBD structures of these sub-variants and a comparison with the known complex structures, we found that R346T substitution in the RBD enhanced ACE2 binding upon an interaction with the residue R493, but not Q493, via a mechanism involving long-range conformation changes. Furthermore, we found that R493Q and F486V exert a balanced impact, through which immune evasion capability was somewhat compromised to achieve an optimal receptor binding. We propose a "two-steps-forward and one-step-backward" model to describe such a compromise between receptor binding affinity and immune evasion during RBD evolution of Omicron sub-variants. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_37471.map.gz emd_37471.map.gz | 56.9 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-37471-v30.xml emd-37471-v30.xml emd-37471.xml emd-37471.xml | 20.3 KB 20.3 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_37471.png emd_37471.png | 52.1 KB | ||
| Filedesc metadata |  emd-37471.cif.gz emd-37471.cif.gz | 6.8 KB | ||
| その他 |  emd_37471_half_map_1.map.gz emd_37471_half_map_1.map.gz emd_37471_half_map_2.map.gz emd_37471_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37471 http://ftp.pdbj.org/pub/emdb/structures/EMD-37471 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37471 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37471 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_37471_validation.pdf.gz emd_37471_validation.pdf.gz | 677.4 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_37471_full_validation.pdf.gz emd_37471_full_validation.pdf.gz | 677 KB | 表示 | |
| XML形式データ |  emd_37471_validation.xml.gz emd_37471_validation.xml.gz | 12.1 KB | 表示 | |
| CIF形式データ |  emd_37471_validation.cif.gz emd_37471_validation.cif.gz | 14 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37471 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37471 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37471 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37471 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8we4MC  8wdrC  8wdsC  8wdyC  8wdzC  8we0C  8we1C M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_37471.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_37471.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
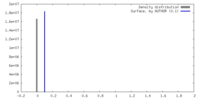
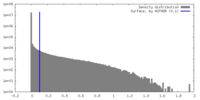
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: #2
| ファイル | emd_37471_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
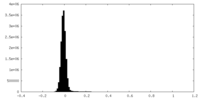
| 密度ヒストグラム |
-ハーフマップ: #1
| ファイル | emd_37471_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
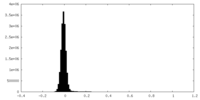
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
+全体 : SARS-CoV-2 Omicron XBB.1.5 RBD complexed with human ACE2 and Fab S304
+超分子 #1: SARS-CoV-2 Omicron XBB.1.5 RBD complexed with human ACE2 and Fab S304
+超分子 #2: ACE2
+超分子 #3: SARS-CoV-2 Omicron XBB.1.5 RBD
+超分子 #4: FabS304
+分子 #1: Angiotensin-converting enzyme 2
+分子 #2: Spike protein S1
+分子 #3: S304 Fab Heavy Chain
+分子 #4: S304 Fab Light Chain
+分子 #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
+分子 #8: ZINC ION
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 0.2 mg/mL |
|---|---|
| 緩衝液 | pH: 8 |
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.0 µm / 最小 デフォーカス(公称値): 1.0 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
- 画像解析
画像解析
| 初期モデル | モデルのタイプ: PDB ENTRY PDBモデル - PDB ID: |
|---|---|
| 最終 再構成 | 解像度のタイプ: BY AUTHOR / 解像度: 2.91 Å / 解像度の算出法: FSC 0.143 CUT-OFF / 使用した粒子像数: 244183 |
| 初期 角度割当 | タイプ: MAXIMUM LIKELIHOOD |
| 最終 角度割当 | タイプ: MAXIMUM LIKELIHOOD |
 ムービー
ムービー コントローラー
コントローラー













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)