[English] 日本語
 Yorodumi
Yorodumi- EMDB-37261: Cryo-EM map of Myosin VI in the autoinhibited state (with SAH-ext... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of Myosin VI in the autoinhibited state (with SAH-extension density) | |||||||||
 Map data Map data | cryo-EM map of autoinhibited MyoVI with SAH-extension | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | myosin VI / autoinhibition / intracellular transport / MOTOR PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.14 Å | |||||||||
 Authors Authors | Niu F / Wei Z | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Autoinhibition and activation of myosin VI revealed by its cryo-EM structure. Authors: Fengfeng Niu / Lingxuan Li / Lei Wang / Jinman Xiao / Shun Xu / Yong Liu / Leishu Lin / Cong Yu / Zhiyi Wei /  Abstract: Myosin VI is the only molecular motor that moves towards the minus end along actin filaments. Numerous cellular processes require myosin VI and tight regulations of the motor's activity. Defects in ...Myosin VI is the only molecular motor that moves towards the minus end along actin filaments. Numerous cellular processes require myosin VI and tight regulations of the motor's activity. Defects in myosin VI activity are known to cause genetic diseases such as deafness and cardiomyopathy. However, the molecular mechanisms underlying the activity regulation of myosin VI remain elusive. Here, we determined the high-resolution cryo-electron microscopic structure of myosin VI in its autoinhibited state. Our structure reveals that autoinhibited myosin VI adopts a compact, monomeric conformation via extensive interactions between the head and tail domains, orchestrated by an elongated single-α-helix region resembling a "spine". This autoinhibited structure effectively blocks cargo binding sites and represses the motor's ATPase activity. Certain cargo adaptors such as GIPC can release multiple inhibitory interactions and promote motor activity, pointing to a cargo-mediated activation of the processive motor. Moreover, our structural findings allow rationalization of disease-associated mutations in myosin VI. Beyond the activity regulation mechanisms of myosin VI, our study also sheds lights on how activities of other myosin motors such as myosin VII and X might be regulated. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37261.map.gz emd_37261.map.gz | 284.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37261-v30.xml emd-37261-v30.xml emd-37261.xml emd-37261.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37261.png emd_37261.png | 71.4 KB | ||
| Filedesc metadata |  emd-37261.cif.gz emd-37261.cif.gz | 5.8 KB | ||
| Others |  emd_37261_half_map_1.map.gz emd_37261_half_map_1.map.gz emd_37261_half_map_2.map.gz emd_37261_half_map_2.map.gz | 301.2 MB 301.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37261 http://ftp.pdbj.org/pub/emdb/structures/EMD-37261 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37261 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37261 | HTTPS FTP |
-Validation report
| Summary document |  emd_37261_validation.pdf.gz emd_37261_validation.pdf.gz | 990 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37261_full_validation.pdf.gz emd_37261_full_validation.pdf.gz | 989.6 KB | Display | |
| Data in XML |  emd_37261_validation.xml.gz emd_37261_validation.xml.gz | 16.9 KB | Display | |
| Data in CIF |  emd_37261_validation.cif.gz emd_37261_validation.cif.gz | 20 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37261 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37261 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37261 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37261 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37261.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37261.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM map of autoinhibited MyoVI with SAH-extension | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.827 Å | ||||||||||||||||||||||||||||||||||||
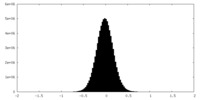
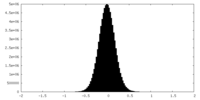
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map B
| File | emd_37261_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half_map_B | ||||||||||||
| Projections & Slices |
| ||||||||||||
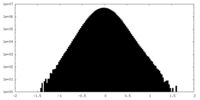
| Density Histograms |
-Half map: half map A
| File | emd_37261_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half_map_A | ||||||||||||
| Projections & Slices |
| ||||||||||||
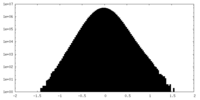
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of Myosin VI and Calmodulin
| Entire | Name: Complex of Myosin VI and Calmodulin |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Myosin VI and Calmodulin
| Supramolecule | Name: Complex of Myosin VI and Calmodulin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Myosin VI
| Supramolecule | Name: Myosin VI / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Calmodulin
| Supramolecule | Name: Calmodulin / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Myosin VI
| Macromolecule | Name: Myosin VI / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEDGKPVWAP HPTDGFQMGN IVDIGPDSLT IEPLNQKGKT FLALINQVFP AEEDSKKDVE DNCSLMYLNE ATLLHNIKVR YSKDRIYTYV ANILIAVNPY FDIPKIYSSE AIKSYQGKSL GTRPPHVFAI ADKAFRDMKV LKMSQSIIVS GESGAGKTEN TKFVLRYLTE ...String: MEDGKPVWAP HPTDGFQMGN IVDIGPDSLT IEPLNQKGKT FLALINQVFP AEEDSKKDVE DNCSLMYLNE ATLLHNIKVR YSKDRIYTYV ANILIAVNPY FDIPKIYSSE AIKSYQGKSL GTRPPHVFAI ADKAFRDMKV LKMSQSIIVS GESGAGKTEN TKFVLRYLTE SYGTGQDIDD RIVEANPLLE AFGNAKTVRN NNSSRFGKFV EIHFNEKSSV VGGFVSHYLL EKSRICVQGK EERNYHIFYR LCAGASEDIR EKLHLSSPDN FRYLNRGCTR YFANKETDKQ ILQNRKSPEY LKAGSMKDPL LDDHGDFIRM CTAMKKIGLD DEEKLDLFRV VAGVLHLGNI DFEEAGSTSG GCNLKNKSAQ SLEYCAELLG LDQDDLRVSL TTRVMLTTAG GTKGTVIKVP LKVEQANNAR DALAKTVYSH LFDHVVNRVN QCFPFETSSY FIGVLDIAGF EYFEHNSFEQ FCINYCNEKL QQFFNERILK EEQELYQKEG LGVNEVHYVD NQDCIDLIEA KLVGILDILD EENRLPQPSD QHFTSAVHQK HKDHFRLTIP RKSKLAVHRN IRDDEGFIIR HFAGAVCYET TQFVEKNNDA LHMSLESLIC ESRDKFIREL FESSTNNNKD TKQKAGKLSF ISVGNKFKTQ LNLLLDKLRS TGASFIRCIK PNLKMTSHHF EGAQILSQLQ CSGMVSVLDL MQGGYPSRAS FHELYNMYKK YMPDKLARLD PRLFCKALFK ALGLNENDYK FGLTKVFFRP GKFAEFDQIM KSDPDHLAEL VKRVNHWLTC SRWKKVQWCS LSVIKLKNKI KYRAEACIKM QKTIRMWLCK RRHKPRIDGL VKVGTLKKRL DKFNEVVSVL KDGKPEMNKQ IKNLEISIDT LMAKIKSTMM TQEQIQKEYD ALVKSSEELL SALQKKKQQE EEAERLRRIQ EEMEKERKRR EEDEKRRRKE EEERRMKLEM EAKRKQEEEE RKKREDDEKR IQAEVEAQLA RQKEEESQQQ AVLEQERRDR ELALRIAQSE AELISDEAQA DLALRRNDGT RPKMTPEQMA KEMSEFLSRG PAVLATKAAA GTKKYDLSKW KYAELRDTIN TSCDIELLAA CREEFHRRLK VYHAWKSKNK KRNTETEQRA PKSVTDYDFA PFLNNSPQQN PAAQIPARQR EIEMNRQQRF FRIPFIRPAD QYKDPQSKKK GWWYAHFDGP WIARQMELHP DKPPILLVAG KDDMEMCELN LEETGLTRKR GAEILPRQFE EIWERCGGIQ YLQNAIESRQ ARPTYATAML QSLLK |
-Macromolecule #2: Calmodulin
| Macromolecule | Name: Calmodulin / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MADQLTEEQI AEFKEAFSLF DKDGDGTITT KELGTVMRSL GQNPTEAELQ DMINEVDADG NGTIDFPEFL TMMARKMKDT DSEEEIREAF RVFDKDGNGY ISAAELRHVM TNLGEKLTDE EVDEMIREAD IDGDGQVNYE EFVQMMTAK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 2.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)