+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bacterial serine protease | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Seine protease / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   | |||||||||
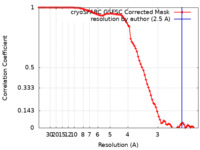
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Lee I-G / Jeon H | |||||||||
| Funding support |  Korea, Republic Of, 1 items Korea, Republic Of, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure of bacterial serine protease Authors: Lee I-G / Jeon H | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37257.map.gz emd_37257.map.gz | 89.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37257-v30.xml emd-37257-v30.xml emd-37257.xml emd-37257.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37257_fsc.xml emd_37257_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_37257.png emd_37257.png | 55.6 KB | ||
| Filedesc metadata |  emd-37257.cif.gz emd-37257.cif.gz | 5.9 KB | ||
| Others |  emd_37257_half_map_1.map.gz emd_37257_half_map_1.map.gz emd_37257_half_map_2.map.gz emd_37257_half_map_2.map.gz | 165.4 MB 165.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37257 http://ftp.pdbj.org/pub/emdb/structures/EMD-37257 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37257 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37257 | HTTPS FTP |
-Validation report
| Summary document |  emd_37257_validation.pdf.gz emd_37257_validation.pdf.gz | 888.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37257_full_validation.pdf.gz emd_37257_full_validation.pdf.gz | 888.3 KB | Display | |
| Data in XML |  emd_37257_validation.xml.gz emd_37257_validation.xml.gz | 19.3 KB | Display | |
| Data in CIF |  emd_37257_validation.cif.gz emd_37257_validation.cif.gz | 24.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37257 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37257 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37257 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37257 | HTTPS FTP |
-Related structure data
| Related structure data |  8kicMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_37257.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37257.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
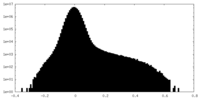
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.848 Å | ||||||||||||||||||||||||||||||||||||
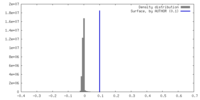
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37257_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
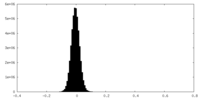
| Density Histograms |
-Half map: #1
| File | emd_37257_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
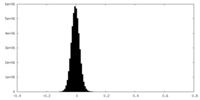
| Density Histograms |
- Sample components
Sample components
-Entire : Bacterial serine protease-lysozyme complex
| Entire | Name: Bacterial serine protease-lysozyme complex |
|---|---|
| Components |
|
-Supramolecule #1: Bacterial serine protease-lysozyme complex
| Supramolecule | Name: Bacterial serine protease-lysozyme complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: peptidase Do
| Macromolecule | Name: peptidase Do / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.361047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKKQTQLLSA LALSVGLTLS ASFQAVASIP GQVADQAPLP SLAPMLEKVL PAVVSVRVEG TASQGQKIPE EFKKFFGDDL PDQPAQPFE GLGSGVIINA SKGYVLTNNH VINQAQKISI QLNDGREFDA KLIGSDDQSD IALLQIQNPS KLTQIAIADS D KLRVGDFA ...String: MKKQTQLLSA LALSVGLTLS ASFQAVASIP GQVADQAPLP SLAPMLEKVL PAVVSVRVEG TASQGQKIPE EFKKFFGDDL PDQPAQPFE GLGSGVIINA SKGYVLTNNH VINQAQKISI QLNDGREFDA KLIGSDDQSD IALLQIQNPS KLTQIAIADS D KLRVGDFA VAVGNPFGLG QTATSGIVSA LGRSGLNLEG LENFIQTDAS INRGNAGGAL LNLNGELIGI NTAILAPGGG SV GIGFAIP SNMARTLAQQ LIDFGEIKRG LLGIKGTEMS ADIAKAFNLD VQRGAFVSEV LPGSGSAKAG VKAGDIITSL NGK PLNSFA ELRSRIATTE PGTKVKLGLL RNGKPLEVEV TLDTSTSSSA SAEMITPALE GATLSDGQLK DGGKGIKIDE VVKG SPAAQ AGLQKDDVII GVNRDRVNSI AEMRKVLAAK PAIIALQIVR GNESIYLLMR LWHHHHHH UniProtKB: peptidase Do |
-Macromolecule #2: Lysozyme fragment (unknown sequence)
| Macromolecule | Name: Lysozyme fragment (unknown sequence) / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 698.854 Da |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #3: Lysozyme fragment (unknown sequence)
| Macromolecule | Name: Lysozyme fragment (unknown sequence) / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 443.539 Da |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #4: Lysozyme fragment (unknown sequence)
| Macromolecule | Name: Lysozyme fragment (unknown sequence) / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 613.749 Da |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 67.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.7 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)