+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of HSV-1 gB with D48 Fab complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HSV-1 gB / fab / neutralizing antibody / complex / Cryo-EM / VIRAL PROTEIN/IMMUNE SYSTEM / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell Golgi membrane / host cell endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / virion membrane / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human herpesvirus 1 (strain KOS) Human herpesvirus 1 (strain KOS) | |||||||||
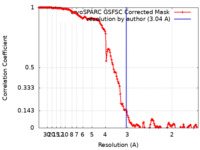
| Method | single particle reconstruction / cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Yang J / Sun C / Fang X / Zeng M / Liu Z | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: hlife / Year: 2023 Journal: hlife / Year: 2023Title: The structure of HSV-1 gB bound to a potent neutralizing antibody reveals a conservative antigenic domain across herpesviruses Authors: Sun C / Yang JW / Xie C / Fang XY / Bu GL / Zhao GX / Dai DL / Liu Z / Zeng MS | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37203.map.gz emd_37203.map.gz | 283.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37203-v30.xml emd-37203-v30.xml emd-37203.xml emd-37203.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37203_fsc.xml emd_37203_fsc.xml | 14.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_37203.png emd_37203.png | 24.1 KB | ||
| Filedesc metadata |  emd-37203.cif.gz emd-37203.cif.gz | 6.3 KB | ||
| Others |  emd_37203_half_map_1.map.gz emd_37203_half_map_1.map.gz emd_37203_half_map_2.map.gz emd_37203_half_map_2.map.gz | 301.9 MB 301.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37203 http://ftp.pdbj.org/pub/emdb/structures/EMD-37203 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37203 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37203 | HTTPS FTP |
-Related structure data
| Related structure data |  8kfaMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_37203.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37203.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.827 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37203_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_37203_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of HSV-1 gB with D48 Fab complex
| Entire | Name: Cryo-EM structure of HSV-1 gB with D48 Fab complex |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of HSV-1 gB with D48 Fab complex
| Supramolecule | Name: Cryo-EM structure of HSV-1 gB with D48 Fab complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: D48 heavy chain
| Macromolecule | Name: D48 heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.742861 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVETGGG VVRPGRSLRL SCTTSGFSFS GSAMHWVRQA PGKGLEWVAV ISHDGNIIQY HDSVKGRFTI SRDNSKNVLL LQMNSLRVD DTAMYYCARD VWLLPATISY AFDFWGQGTM VTVSSASTKG PSVFPLAPSS KSTSGGTAAL GCLVKDYFPE P VTVSWNSG ...String: EVQLVETGGG VVRPGRSLRL SCTTSGFSFS GSAMHWVRQA PGKGLEWVAV ISHDGNIIQY HDSVKGRFTI SRDNSKNVLL LQMNSLRVD DTAMYYCARD VWLLPATISY AFDFWGQGTM VTVSSASTKG PSVFPLAPSS KSTSGGTAAL GCLVKDYFPE P VTVSWNSG ALTSGVHTFP AVLQSSGLYS LSSVVTVPSS SLGTQTYICN VNHKPSNTKV DKRVGSHHHH HH |
-Macromolecule #2: D48 light chain
| Macromolecule | Name: D48 light chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.476164 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VIWMTQSPPS LSASIGDTVT ITCRASQGIS NSIAWYQRRP GKAPELLVYA AYRLQSGVPS RLSGSGSGAE YTLTIKNMQP EDFATYYCQ QYYDNPLTFG GGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: VIWMTQSPPS LSASIGDTVT ITCRASQGIS NSIAWYQRRP GKAPELLVYA AYRLQSGVPS RLSGSGSGAE YTLTIKNMQP EDFATYYCQ QYYDNPLTFG GGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #3: Envelope glycoprotein B
| Macromolecule | Name: Envelope glycoprotein B / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 1 (strain KOS) Human herpesvirus 1 (strain KOS) |
| Molecular weight | Theoretical: 70.337844 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ANFYVCPPPT GATVVQFEQP RRCPTRPEGQ NYTEGIAVVF KENIAPYKFK ATMYYKDVTV SQVWFGHRYS QFMGIFEDRA PVPFEEVID KINAKGVCRS TAKYVRNNLE TTAFHRDDHE TDMELKPANA ATRTSRGWHT TDLKYNPSRV EAFHRYGTTV N CIVEEVDA ...String: ANFYVCPPPT GATVVQFEQP RRCPTRPEGQ NYTEGIAVVF KENIAPYKFK ATMYYKDVTV SQVWFGHRYS QFMGIFEDRA PVPFEEVID KINAKGVCRS TAKYVRNNLE TTAFHRDDHE TDMELKPANA ATRTSRGWHT TDLKYNPSRV EAFHRYGTTV N CIVEEVDA RSVYPYDEFV LATGDFVYMS PFYGYREGSH TEHTTYAADR FKQVDGFYAR DLTTKARATA PTTRNLLTTP KF TVAWDWV PKRPSVCTMT KWQEVDEMLR SEYGGSFRFS SDAISTTFTT NLTEYPLSRV DLGDCIGKDA RDAMDRIFAR RYN ATHIKV GQPQYYQANG GFLIAYQPLL SNTLAELYVR EHLREQSRKP PNPTPPPPGA SANASVERIK TTSSIEFARL QFTY NHIQR HVNDMLGRVA IAWCELQNHE LTLWNEARKL NPNAIASVTV GRRVSARMLG DVMAVSTCVP VAADNVIVQN SMRIS SRPG ACYSRPLVSF RYEDQGPLVE GQLGENNELR LTRDAIEPCT VGHRRYFTFG GGYVYFEEYA YSHQLSRADI TTVSTF IDL NITMLEDHEF VPLEVYTRHE IKDSGLLDYT EVQRRNQLHD LRFADIDTVI HA UniProtKB: Envelope glycoprotein B |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.56 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)