+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human SIDT1 bound to cholesterol | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Membrane protein | ||||||||||||
| Function / homology | RNA transmembrane transporter activity / SID1 transmembrane family / dsRNA-gated channel SID-1 / RNA transport / cholesterol binding / double-stranded RNA binding / lysosome / plasma membrane / SID1 transmembrane family member 1 Function and homology information Function and homology information | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.77 Å | ||||||||||||
 Authors Authors | Hirano Y / Ohto U / Shimizu T | ||||||||||||
| Funding support |  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2024 Journal: Commun Biol / Year: 2024Title: Cryo-EM analysis reveals human SID-1 transmembrane family member 1 dynamics underlying lipid hydrolytic activity. Authors: Yoshinori Hirano / Umeharu Ohto / Ikuyo Ichi / Ryota Sato / Kensuke Miyake / Toshiyuki Shimizu /  Abstract: Two mammalian homologs of systemic RNA interference defective protein 1 (SID-1) (SIDT1/2) are suggested to function as double-stranded RNA (dsRNA) transporters for extracellular dsRNA uptake or for ...Two mammalian homologs of systemic RNA interference defective protein 1 (SID-1) (SIDT1/2) are suggested to function as double-stranded RNA (dsRNA) transporters for extracellular dsRNA uptake or for release of incorporated dsRNA from lysosome to cytoplasm. SIDT1/2 is also suggested to be involved in cholesterol transport and lipid metabolism. Here, we determine the cryo-electron microscopy structures of human SIDT1, homodimer in a side-by-side arrangement, with two distinct conformations, the cholesterol-bound form and the unbound form. Our structures reveal that the membrane-spanning region of SIDT1 harbors conserved histidine and aspartate residues coordinating to putative zinc ion, in a structurally similar manner to alkaline ceramidases or adiponectin receptors that require zinc for ceramidase activity. We identify that SIDT1 has a ceramidase activity that is attenuated by cholesterol binding. Observations from two structures suggest that cholesterol molecules serve as allosteric regulator that binds the transmembrane region of SIDT1 and induces the conformation change and the reorientation of the catalytic residues. This study represents a contribution to the elucidation of the cholesterol-mediated mechanisms of lipid hydrolytic activity and RNA transport in the SID-1 family proteins. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37112.map.gz emd_37112.map.gz | 22.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37112-v30.xml emd-37112-v30.xml emd-37112.xml emd-37112.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37112.png emd_37112.png | 98.2 KB | ||
| Filedesc metadata |  emd-37112.cif.gz emd-37112.cif.gz | 6 KB | ||
| Others |  emd_37112_half_map_1.map.gz emd_37112_half_map_1.map.gz emd_37112_half_map_2.map.gz emd_37112_half_map_2.map.gz | 22.5 MB 22.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37112 http://ftp.pdbj.org/pub/emdb/structures/EMD-37112 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37112 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37112 | HTTPS FTP |
-Validation report
| Summary document |  emd_37112_validation.pdf.gz emd_37112_validation.pdf.gz | 817.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37112_full_validation.pdf.gz emd_37112_full_validation.pdf.gz | 817 KB | Display | |
| Data in XML |  emd_37112_validation.xml.gz emd_37112_validation.xml.gz | 10.5 KB | Display | |
| Data in CIF |  emd_37112_validation.cif.gz emd_37112_validation.cif.gz | 12.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37112 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37112 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37112 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37112 | HTTPS FTP |
-Related structure data
| Related structure data |  8kcwMC  8kcxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_37112.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37112.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.245 Å | ||||||||||||||||||||||||||||||||||||
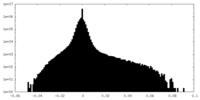
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37112_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
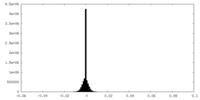
| Density Histograms |
-Half map: #1
| File | emd_37112_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
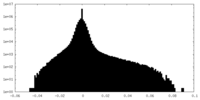
| Density Histograms |
- Sample components
Sample components
-Entire : homo-dimer of SIDT1
| Entire | Name: homo-dimer of SIDT1 |
|---|---|
| Components |
|
-Supramolecule #1: homo-dimer of SIDT1
| Supramolecule | Name: homo-dimer of SIDT1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: SID1 transmembrane family member 1
| Macromolecule | Name: SID1 transmembrane family member 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 96.877312 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRGCLRLALL CALPWLLLAA SPGHPAKSPR QPPAPRRDPF DAARGADFDH VYSGVVNLST ENIYSFNYTS QPDQVTAVRV YVNSSSENL NYPVLVVVRQ QKEVLSWQVP LLFQGLYQRS YNYQEVSRTL CPSEATNETG PLQQLIFVDV ASMAPLGAQY K LLVTKLKH ...String: MRGCLRLALL CALPWLLLAA SPGHPAKSPR QPPAPRRDPF DAARGADFDH VYSGVVNLST ENIYSFNYTS QPDQVTAVRV YVNSSSENL NYPVLVVVRQ QKEVLSWQVP LLFQGLYQRS YNYQEVSRTL CPSEATNETG PLQQLIFVDV ASMAPLGAQY K LLVTKLKH FQLRTNVAFH FTASPSQPQY FLYKFPKDVD SVIIKVVSEM AYPCSVVSVQ NIMCPVYDLD HNVEFNGVYQ SM TKKAAIT LQKKDFPGEQ FFVVFVIKPE DYACGGSFFI QEKENQTWNL QRKKNLEVTI VPSIKESVYV KSSLFSVFIF LSF YLGCLL VGFVHYLRFQ RKSIDGSFGS NDGSGNMVAS HPIAASTPEG SNYGTIDESS SSPGRQMSSS DGGPPGQSDT DSSV EESDF DTMPDIESDK NIIRTKMFLY LSDLSRKDRR IVSKKYKIYF WNIITIAVFY ALPVIQLVIT YQTVVNVTGN QDICY YNFL CAHPLGVLSA FNNILSNLGH VLLGFLFLLI VLRRDILHRR ALEAKDIFAV EYGIPKHFGL FYAMGIALMM EGVLSA CYH VCPNYSNFQF DTSFMYMIAG LCMLKLYQTR HPDINASAYS AYASFAVVIM VTVLGVVFGK NDVWFWVIFS AIHVLAS LA LSTQIYYMGR FKIDLGIFRR AAMVFYTDCI QQCSRPLYMD RMVLLVVGNL VNWSFALFGL IYRPRDFASY MLGIFICN L LLYLAFYIIM KLRSSEKVLP VPLFCIVATA VMWAAALYFF FQNLSSWEGT PAESREKNRE CILLDFFDDH DIWHFLSAT ALFFSFLVLL TLDDDLDVVR RDQIPVFENL YFQGDYKDDD DKHHHHHHHH UniProtKB: SID1 transmembrane family member 1 |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #5: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 5 / Number of copies: 4 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 6 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 65.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.77 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 59745 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)