+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | the alpha-galactosidase 5 with Cacl2 | |||||||||
 Map data Map data | this is map of alpha-galactosidases with cacl2. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | alpha-Galactosidase / HYDROLASE / Cacl2 | |||||||||
| Function / homology |  Function and homology information Function and homology informationalpha-galactosidase / alpha-galactosidase activity / carbohydrate catabolic process Similarity search - Function | |||||||||
| Biological species |  Blautia pseudococcoides (bacteria) Blautia pseudococcoides (bacteria) | |||||||||
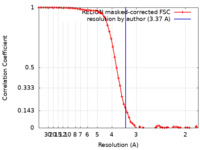
| Method | single particle reconstruction / cryo EM / Resolution: 3.37 Å | |||||||||
 Authors Authors | Li YW / Ru YX | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Chem / Year: 2024 Journal: Commun Chem / Year: 2024Title: Activity-based metaproteomics driven discovery and enzymological characterization of potential α-galactosidases in the mouse gut microbiome. Authors: Jianbing Jiang / Diana Czuchry / Yanxia Ru / Huipai Peng / Junfeng Shen / Teng Wang / Wenjuan Zhao / Weihua Chen / Sen-Fang Sui / Yaowang Li / Nan Li /  Abstract: The gut microbiota offers an extensive resource of enzymes, but many remain uncharacterized. To distinguish the activities of similar annotated proteins and mine the potentially applicable ones in ...The gut microbiota offers an extensive resource of enzymes, but many remain uncharacterized. To distinguish the activities of similar annotated proteins and mine the potentially applicable ones in the microbiome, we applied an effective Activity-Based Metaproteomics (ABMP) strategy using a specific activity-based probe (ABP) to screen the entire gut microbiome for directly discovering active enzymes and their potential applications, not for exploring host-microbiome interactions. By using an activity-based cyclophellitol aziridine probe specific to α-galactosidases (AGAL), we successfully identified and characterized several gut microbiota enzymes possessing AGAL activities. Cryo-electron microscopy analysis of a newly characterized enzyme (AGLA5) revealed the covalent binding conformations between the AGAL5 active site and the cyclophellitol aziridine ABP, which could provide insights into the enzyme's catalytic mechanism. The four newly characterized AGALs have diverse potential activities, including raffinose family oligosaccharides (RFOs) hydrolysis and enzymatic blood group transformation. Collectively, we present a ABMP platform that facilitates gut microbiota AGALs discovery, biochemical activity annotations and potential industrial or biopharmaceutical applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36943.map.gz emd_36943.map.gz | 20.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36943-v30.xml emd-36943-v30.xml emd-36943.xml emd-36943.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36943_fsc.xml emd_36943_fsc.xml | 6.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_36943.png emd_36943.png | 40.6 KB | ||
| Filedesc metadata |  emd-36943.cif.gz emd-36943.cif.gz | 6.3 KB | ||
| Others |  emd_36943_half_map_1.map.gz emd_36943_half_map_1.map.gz emd_36943_half_map_2.map.gz emd_36943_half_map_2.map.gz | 17 MB 17 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36943 http://ftp.pdbj.org/pub/emdb/structures/EMD-36943 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36943 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36943 | HTTPS FTP |
-Validation report
| Summary document |  emd_36943_validation.pdf.gz emd_36943_validation.pdf.gz | 925.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36943_full_validation.pdf.gz emd_36943_full_validation.pdf.gz | 925.2 KB | Display | |
| Data in XML |  emd_36943_validation.xml.gz emd_36943_validation.xml.gz | 11.9 KB | Display | |
| Data in CIF |  emd_36943_validation.cif.gz emd_36943_validation.cif.gz | 16.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36943 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36943 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36943 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36943 | HTTPS FTP |
-Related structure data
| Related structure data |  8k7uMC  8k1aC  8k7vC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36943.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36943.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | this is map of alpha-galactosidases with cacl2. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.92 Å | ||||||||||||||||||||||||||||||||||||
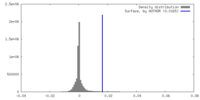
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: this is half map1.
| File | emd_36943_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | this is half map1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
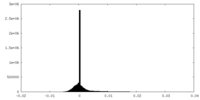
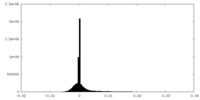
| Density Histograms |
-Half map: this is half map2.
| File | emd_36943_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | this is half map2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
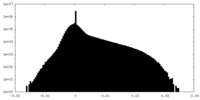
| Density Histograms |
- Sample components
Sample components
-Entire : The tetramer alpha-galactosidase 5 protein with Cacl2
| Entire | Name: The tetramer alpha-galactosidase 5 protein with Cacl2 |
|---|---|
| Components |
|
-Supramolecule #1: The tetramer alpha-galactosidase 5 protein with Cacl2
| Supramolecule | Name: The tetramer alpha-galactosidase 5 protein with Cacl2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Blautia pseudococcoides (bacteria) Blautia pseudococcoides (bacteria) |
-Macromolecule #1: Alpha-galactosidase
| Macromolecule | Name: Alpha-galactosidase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Blautia pseudococcoides (bacteria) Blautia pseudococcoides (bacteria) |
| Molecular weight | Theoretical: 88.017266 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAVIFHEKTK EFHIFNREVS YLMRIMENGQ LENLYYGKVI RDKEDFGYLH EEAMRSQMSV CIPEPGILSM QYTRQEYPVY GTGDYRSPA LTVLQENGSR LVDFSYVSHE IYKGKKGIPP LPSTYAESED EAETLEVTLH DQVTDTDLVL TYTIYEDYPV I TRNARFEQ ...String: MAVIFHEKTK EFHIFNREVS YLMRIMENGQ LENLYYGKVI RDKEDFGYLH EEAMRSQMSV CIPEPGILSM QYTRQEYPVY GTGDYRSPA LTVLQENGSR LVDFSYVSHE IYKGKKGIPP LPSTYAESED EAETLEVTLH DQVTDTDLVL TYTIYEDYPV I TRNARFEQ KGEQKIVLER AMSASVEFLD MDYELVQLSG AWSRERYVKN RKLEMGIQSV HSLNGTCGGA EHNPFIALKR PQ TTENQGE VYGFSLVYSG NFLAQAEVST FDMTRVMLGI NPEDFSWELN QGESFQTPEV VMVYSDRGLN KMSQAYHRLY RTR LMRVTW RDKARPILLN NWEATYFDFN EEKILKIAEK AKEAGVELFV LDDGWFGARN DDYRGLGDWY VNLEKLPDGI AGLS RKVEA LGLKFGLWVE LEMVNKDSDL YRAHPDWLIG APDRFESHAR HQHVLDFSRK EVVDYIYKMI AKVLRESSIS YIKWD MNRY MTEPYSRGAD ASQQGKVMHK YILGVYDLYT RLTTEFPEIL FESCASGGAR FDPAMLYFAP QTWTSDDTDA SERTKI QYG TSYVYPVVSM GSHVSAVPNH QMHRMTPIET RANVAYFGTF GYELDLNLLS EAELESVKKQ IAFMKEYREL IQVDGDF YR LLSPFEGNET AWMVVAQDKS RAVAAFYQRM NKVNASWIRF KLQGLDAGTL YEVSCDMAPS ASYDESLAKI YGIQTEEN M VKTYRAYGDE LMQVGIPIDR EDLNKKGGDF ASLLYTLKKV TD UniProtKB: Alpha-galactosidase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



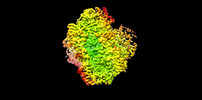


 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)