+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryo-EM map of close TIEA-TEC complex | |||||||||
 Map data Map data | The cryo-EM map of close TIEA-TIC complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacteriophage T4 / RNA polymerase / transcription / inhibitor / elongation accelerator / complex / TRANSCRIPTION/DNA/RNA / TRANSCRIPTION-DNA-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA polymerase complex / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / bacterial-type flagellum assembly / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / bacterial-type flagellum-dependent cell motility / nitrate assimilation / regulation of DNA-templated transcription elongation ...RNA polymerase complex / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / bacterial-type flagellum assembly / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / bacterial-type flagellum-dependent cell motility / nitrate assimilation / regulation of DNA-templated transcription elongation / transcription elongation factor complex / transcription antitermination / cell motility / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / response to heat / protein-containing complex assembly / intracellular iron ion homeostasis / DNA replication / protein dimerization activity / response to antibiotic / magnesium ion binding / DNA binding / zinc ion binding / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Escherichia phage T4 (virus) Escherichia phage T4 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.15 Å | |||||||||
 Authors Authors | Zhang KN / Liu Y / Chen M / Wang Y / Lin W / Li M / Zhang X / Gao Y / Gong Q / Chen H ...Zhang KN / Liu Y / Chen M / Wang Y / Lin W / Li M / Zhang X / Gao Y / Gong Q / Chen H / Steve M / Li S / Zhang K / Liu B | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: TIEA inhibits Sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error Authors: Zhang KN / Liu Y / Chen M / Wang Y / Lin W / Li M / Zhang X / Gao Y / Gong Q / Chen H / Steve M / Li S / Zhang K / Liu B | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36897.map.gz emd_36897.map.gz | 136.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36897-v30.xml emd-36897-v30.xml emd-36897.xml emd-36897.xml | 29.3 KB 29.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36897.png emd_36897.png | 78.3 KB | ||
| Filedesc metadata |  emd-36897.cif.gz emd-36897.cif.gz | 9 KB | ||
| Others |  emd_36897_additional_1.map.gz emd_36897_additional_1.map.gz emd_36897_half_map_1.map.gz emd_36897_half_map_1.map.gz emd_36897_half_map_2.map.gz emd_36897_half_map_2.map.gz | 72.2 MB 134.5 MB 134.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36897 http://ftp.pdbj.org/pub/emdb/structures/EMD-36897 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36897 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36897 | HTTPS FTP |
-Validation report
| Summary document |  emd_36897_validation.pdf.gz emd_36897_validation.pdf.gz | 1002.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36897_full_validation.pdf.gz emd_36897_full_validation.pdf.gz | 1002.4 KB | Display | |
| Data in XML |  emd_36897_validation.xml.gz emd_36897_validation.xml.gz | 14.3 KB | Display | |
| Data in CIF |  emd_36897_validation.cif.gz emd_36897_validation.cif.gz | 17 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36897 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36897 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36897 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36897 | HTTPS FTP |
-Related structure data
| Related structure data |  8k58MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36897.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36897.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The cryo-EM map of close TIEA-TIC complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
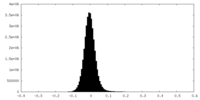
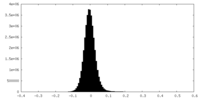
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: The cryo-EM map of close TIEA-TIC complex
| File | emd_36897_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The cryo-EM map of close TIEA-TIC complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
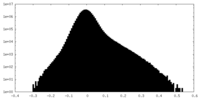
| Density Histograms |
-Half map: The cryo-EM map of close TIEA-TIC complex
| File | emd_36897_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The cryo-EM map of close TIEA-TIC complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: The cryo-EM map of close TIEA-TIC complex
| File | emd_36897_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The cryo-EM map of close TIEA-TIC complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The cryo-EM map of close TIEA-TEC complex
| Entire | Name: The cryo-EM map of close TIEA-TEC complex |
|---|---|
| Components |
|
-Supramolecule #1: The cryo-EM map of close TIEA-TEC complex
| Supramolecule | Name: The cryo-EM map of close TIEA-TEC complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: DNA-directed RNA polymerase subunit beta
| Macromolecule | Name: DNA-directed RNA polymerase subunit beta / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 150.590547 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: YSYTEKKRIR KDFGKRPQVL DVPYLLSIQL DSFQKFIEQD PEGQYGLEAA FRSVFPIQSY SGNSELQYVS YRLGEPVFDV QECQIRGVT YSAPLRVKLR LVIYEREAPE GTVKDIKEQE VYMGEIPLMT DNGTFVINGT ERVIVSQLHR SPGVFFDSDK G KTHSSGKV ...String: YSYTEKKRIR KDFGKRPQVL DVPYLLSIQL DSFQKFIEQD PEGQYGLEAA FRSVFPIQSY SGNSELQYVS YRLGEPVFDV QECQIRGVT YSAPLRVKLR LVIYEREAPE GTVKDIKEQE VYMGEIPLMT DNGTFVINGT ERVIVSQLHR SPGVFFDSDK G KTHSSGKV LYNARIIPYR GSWLDFEFDP KDNLFVRIDR RRKLPATIIL RALNYTTEQI LDLFFEKVIF EIRDNKLQME LV PERLRGE TASFDIEANG KVYVEKGRRI TARHIRQLEK DDVKLIEVPV EYIAGKVVAK DYIDESTGEL ICAANMELSL DLL AKLSQS GHKRIETLFT NDLDHGPYIS ETLRVDPTND RLSALVEIYR MMRPGEPPTR EAAESLFENL FFSEDRYDLS AVGR MKFNR SLLREEIEGS GILSKDDIID VMKKLIDIRN GKGEVDDIDH LGNRRIRSVG EMAENQFRVG LVRVERAVKE RLSLG DLDT LMPQDMINAK PISAAVKEFF GSSQLSQFMD QNNPLSEITH KRRISALGPG GLTRERAGFE VRDVHPTHYG RVCPIE TPE GPNIGLINSL SVYAQTNEYG FLETPYRKVT DGVVTDEIHY LSAIEEGNYV IAQANSNLDE EGHFVEDLVT CRSKGES SL FSRDQVDYMD VSTQQVVSVG ASLIPFLEHD DANRALMGAN MQRQAVPTLR ADKPLVGTGM ERAVAVDSGV TAVAKRGG V VQYVDASRIV IKVNEDEMYP GEAGIDIYNL TKYTRSNQNT CINQMPCVSL GEPVERGDVL ADGPSTDLGE LALGQNMRV AFMPWNGYNF EDSILVSERV VQEDRFTTIH IQELACVSRD TKLGPEEITA DIPNVGEAAL SKLDESGIVY IGAEVTGGDI LVGKVTPKG ETQLTPEEKL LRAIFGEKAS DVKDSSLRVP NGVSGTVIDV QVFTRDGVEK DKRALEIEEM QLKQAKKDLS E ELQILEAG LFSRIRAVLV AGGVEAEKLD KLPRDRWLEL GLTDEEKQNQ LEQLAEQYDE LKHEFEKKLE AKRRKITQGD DL APGVLKI VKVYLAVKRR IQPGDKMAGR HGNKGVISKI NPIEDMPYDE NGTPVDIVLN PLGVPSRMNI GQILETHLGM AAK GIGDKI NAMLKQQQEV AKLREFIQRA YDLGADVRQK VDLSTFSDEE VMRLAENLRK GMPIATPVFD GAKEAEIKEL LKLG DLPTS GQIRLYDGRT GEQFERPVTV GYMYMLKLNH LVDDKMHARS TGSYSLVTQQ PLGGKAQFGG QRFGEMEVWA LEAYG AAYT LQEMLTVKSD DVNGRTKMYK NIVDGNHQME PGMPESFNVL LKEIRSLGIN IELEDE UniProtKB: DNA-directed RNA polymerase subunit beta |
-Macromolecule #2: DNA-directed RNA polymerase subunit beta'
| Macromolecule | Name: DNA-directed RNA polymerase subunit beta' / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 150.865766 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TEEFDAIKIA LASPDMIRSW SFGEVKKPET INYRTFKPER DGLFCARIFG PVKDYECLCG KYKRLKHRGV ICEKCGVEVT QTKVRRERM GHIELASPTA HIWFLKSLPS RIGLLLDMPL RDIERVLYFE SYVVIEGGMT NLERQQILTE EQYLDALEEF G DEFDAKMG ...String: TEEFDAIKIA LASPDMIRSW SFGEVKKPET INYRTFKPER DGLFCARIFG PVKDYECLCG KYKRLKHRGV ICEKCGVEVT QTKVRRERM GHIELASPTA HIWFLKSLPS RIGLLLDMPL RDIERVLYFE SYVVIEGGMT NLERQQILTE EQYLDALEEF G DEFDAKMG AEAIQALLKS MDLEQECEQL REELNETNSE TKRKKLTKRI KLLEAFVQSG NKPEWMILTV LPVLPPDLRP LV PLDGGRF ATSDLNDLYR RVINRNNRLK RLLDLAAPDI IVRNEKRMLQ EAVDALLDNG RRGRAITGSN KRPLKSLADM IKG KQGRFR QNLLGKRVDY SGRSVITVGP YLRLHQCGLP KKMALELFKP FIYGKLELRG LATTIKAAKK MVEREEAVVW DILD EVIRE HPVLLNRAPT LHRLGIQAFE PVLIEGKAIQ LHPLVCAAYN ADFDGDQMAV HVPLTLEAQL EARALMMSTN NILSP ANGE PIIVPSQDVV LGLYYMTRDC VNAKGEGMVL TGPKEAERLY RSGLASLHAR VKVRITEYEK DANGELVAKT SLKDTT VGR AILWMIVPKG LPYSIVNQAL GKKAISKMLN TCYRILGLKP TVIFADQIMY TGFAYAARSG ASVGIDDMVI PEKKHEI IS EAEAEVAEIQ EQFQSGLVTA GERYNKVIDI WAAANDRVSK AMMDNLQTET VINRDGQEEK QVSFNSIYMM ADSGARGS A AQIRQLAGMR GLMAKPDGSI IETPITANFR EGLNVLQYFI STHGARKGLA DTALKTANSG YLTRRLVDVA QDLVVTEDD CGTHEGIMMT PVIEGGDVKE PLRDRVLGRV TAEDVLKPGT ADILVPRNTL LHEQWCDLLE ENSVDAVKVR SVVSCDTDFG VCAHCYGRD LARGHIINKG EAIGVIAAQS IGEPGTQLTM RTFHIGGAAS RAAAESSIQV KNKGSIKLSN VKSVVNSSGK L VITSRNTE LKLIDEFGRT KESYKVPYGA VLAKGDGEQV AGGETVANWD PHTMPVITEV SGFVRFTDMI DGQTITRQTD EL TGLSSLV VLDSAERTAG GKDLRPALKI VDAQGNDVLI PGTDMPAQYF LPGKAIVQLE DGVQISSGDT LARIPQESGG TKD ITGGLP RVADLFEARR PKEPAILAEI SGIVSFGKET KGKRRLVITP VDGSDPYEEM IPKWRQLNVF EGERVERGDV ISDG PEAPH DILRLRGVHA VTRYIVNEVQ DVYRLQGVKI NDKHIEVIVR QMLRKATIVN AGSSDFLEGE QVEYSRVKIA NRELE ANGK VGATYSRDLL GITKASLATE SFISAASFQE TTRVLTEAAV AGKRDELRGL KENVIVGRLI PAGTGYAYHQ DRMRRR AAG UniProtKB: DNA-directed RNA polymerase subunit beta' |
-Macromolecule #3: DNA-directed RNA polymerase subunit omega
| Macromolecule | Name: DNA-directed RNA polymerase subunit omega / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.649695 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ARVTVQDAVE KIGNRFDLVL VAARRARQMQ VGGKDPLVPE ENDKTTVIAL REIEEGLINN QILDVRERQE QQEQEA UniProtKB: DNA-directed RNA polymerase subunit omega |
-Macromolecule #4: DNA-directed RNA polymerase subunit alpha
| Macromolecule | Name: DNA-directed RNA polymerase subunit alpha / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.612111 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TEFLKPRLVD IEQVSSTHAK VTLEPLERGF GHTLGNALRR ILLSSMPGCA VTEVEIDGVL HEYSTKEGVQ EDILEILLNL KGLAVRVQG KDEVILTLNK SGIGPVTAAD ITHDGDVEIV KPQHVICHLT DENASISMRI KVQRGRGYVP ASTRIHSEED E RPIGRLLV ...String: TEFLKPRLVD IEQVSSTHAK VTLEPLERGF GHTLGNALRR ILLSSMPGCA VTEVEIDGVL HEYSTKEGVQ EDILEILLNL KGLAVRVQG KDEVILTLNK SGIGPVTAAD ITHDGDVEIV KPQHVICHLT DENASISMRI KVQRGRGYVP ASTRIHSEED E RPIGRLLV DACYSPVERI AYNVEAARVE QRTDLDKLVI EMETNGTIDP EEAIRRAATI LAEQLEAFVD LRD UniProtKB: DNA-directed RNA polymerase subunit alpha |
-Macromolecule #5: 15 kDa RNA polymerase-binding protein
| Macromolecule | Name: 15 kDa RNA polymerase-binding protein / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage T4 (virus) Escherichia phage T4 (virus) |
| Molecular weight | Theoretical: 14.731656 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTKITVNYTV DVKDIQPKHV RSESNPQNQN KIRRAWVLSL SDNAMEVIQN KIKSAPARHA YYEAIDREVS NKWIELMRKH TTESLNAGA KFIMTSCGER LEDDYCGNAD ERLIVAAQIV AETIAADFNR UniProtKB: 15 kDa RNA polymerase-binding protein |
-Macromolecule #6: DNA (29-MER)
| Macromolecule | Name: DNA (29-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.915744 KDa |
| Sequence | String: (DG)(DG)(DG)(DC)(DT)(DA)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DA)(DT)(DG)(DA)(DC)(DG)(DG) (DC)(DG)(DA)(DA)(DT)(DA)(DC)(DC)(DC) |
-Macromolecule #7: DNA (29-MER)
| Macromolecule | Name: DNA (29-MER) / type: dna / ID: 7 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.813646 KDa |
| Sequence | String: (DG)(DG)(DG)(DT)(DA)(DT)(DT)(DC)(DG)(DC) (DC)(DG)(DT)(DG)(DT)(DA)(DC)(DC)(DT)(DC) (DT)(DC)(DC)(DT)(DA)(DG)(DC)(DC)(DC) |
-Macromolecule #8: RNA (5'-R(*CP*GP*GP*AP*GP*AP*GP*GP*UP*A)-3')
| Macromolecule | Name: RNA (5'-R(*CP*GP*GP*AP*GP*AP*GP*GP*UP*A)-3') / type: rna / ID: 8 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 3.280036 KDa |
| Sequence | String: CGGAGAGGUA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 4092 pixel / Digitization - Dimensions - Height: 5760 pixel / Number real images: 8645 / Average exposure time: 3.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)