[English] 日本語
 Yorodumi
Yorodumi- EMDB-36896: Structure of the Dimeric Human CNTN2 Ig 1-6-FNIII 1-2 Domain in a... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Dimeric Human CNTN2 Ig 1-6-FNIII 1-2 Domain in an Asymmetric State | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | neural cell adhesion / CELL ADHESION | |||||||||
| Function / homology |  Function and homology information Function and homology informationestablishment of protein localization to juxtaparanode region of axon / presynaptic membrane organization / regulation of axon diameter / positive regulation of adenosine receptor signaling pathway / L1CAM interactions / regulation of astrocyte differentiation / reduction of food intake in response to dietary excess / clustering of voltage-gated potassium channels / cerebral cortex GABAergic interneuron migration / dendrite self-avoidance ...establishment of protein localization to juxtaparanode region of axon / presynaptic membrane organization / regulation of axon diameter / positive regulation of adenosine receptor signaling pathway / L1CAM interactions / regulation of astrocyte differentiation / reduction of food intake in response to dietary excess / clustering of voltage-gated potassium channels / cerebral cortex GABAergic interneuron migration / dendrite self-avoidance / protein localization to juxtaparanode region of axon / NrCAM interactions / central nervous system myelination / cell-cell adhesion mediator activity / positive regulation of protein processing / G protein-coupled adenosine receptor signaling pathway / axon initial segment / NCAM1 interactions / juxtaparanode region of axon / node of Ranvier / axonal fasciculation / regulation of cell morphogenesis / adult walking behavior / negative regulation of neuron differentiation / fat cell differentiation / homophilic cell-cell adhesion / regulation of neuronal synaptic plasticity / side of membrane / axon guidance / learning / establishment of localization in cell / synapse organization / protein processing / receptor internalization / microtubule cytoskeleton organization / myelin sheath / carbohydrate binding / postsynaptic membrane / cell adhesion / axon / neuronal cell body / synapse / cell surface / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Zhang ZZ | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure of human CNTN2 immunoglobulin domains 1-6 homo-dimer Authors: Zhang ZZ / Shi ZB / Wang DP | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36896.map.gz emd_36896.map.gz | 230.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36896-v30.xml emd-36896-v30.xml emd-36896.xml emd-36896.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36896.png emd_36896.png | 82 KB | ||
| Filedesc metadata |  emd-36896.cif.gz emd-36896.cif.gz | 6.3 KB | ||
| Others |  emd_36896_half_map_1.map.gz emd_36896_half_map_1.map.gz emd_36896_half_map_2.map.gz emd_36896_half_map_2.map.gz | 226.5 MB 226.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36896 http://ftp.pdbj.org/pub/emdb/structures/EMD-36896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36896 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36896 | HTTPS FTP |
-Related structure data
| Related structure data |  8k53MC  8k3jC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36896.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36896.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.92 Å | ||||||||||||||||||||||||||||||||||||
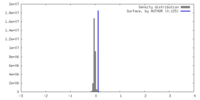
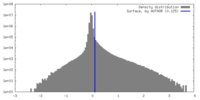
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36896_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
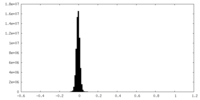
| Density Histograms |
-Half map: #1
| File | emd_36896_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
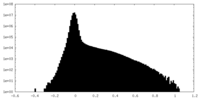
| Density Histograms |
- Sample components
Sample components
-Entire : CNTN2
| Entire | Name: CNTN2 |
|---|---|
| Components |
|
-Supramolecule #1: CNTN2
| Supramolecule | Name: CNTN2 / type: cell / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Contactin-2
| Macromolecule | Name: Contactin-2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 107.585203 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SQTTFGPVFE DQPLSVLFPE ESTEEQVLLA CRARASPPAT YRWKMNGTEM KLEPGSRHQL VGGNLVIMNP TKAQDAGVYQ CLASNPVGT VVSREAILRF GFLQEFSKEE RDPVKAHEGW GVMLPCNPPA HYPGLSYRWL LNEFPNFIPT DGRHFVSQTT G NLYIARTN ...String: SQTTFGPVFE DQPLSVLFPE ESTEEQVLLA CRARASPPAT YRWKMNGTEM KLEPGSRHQL VGGNLVIMNP TKAQDAGVYQ CLASNPVGT VVSREAILRF GFLQEFSKEE RDPVKAHEGW GVMLPCNPPA HYPGLSYRWL LNEFPNFIPT DGRHFVSQTT G NLYIARTN ASDLGNYSCL ATSHMDFSTK SVFSKFAQLN LAAEDTRLFA PSIKARFPAE TYALVGQQVT LECFAFGNPV PR IKWRKVD GSLSPQWTTA EPTLQIPSVS FEDEGTYECE AENSKGRDTV QGRIIVQAQP EWLKVISDTE ADIGSNLRWG CAA AGKPRP TVRWLRNGEP LASQNRVEVL AGDLRFSKLS LEDSGMYQCV AENKHGTIYA SAELAVQALA PDFRLNPVRR LIPA ARGGE ILIPCQPRAA PKAVVLWSKG TEILVNSSRV TVTPDGTLII RNISRSDEGK YTCFAENFMG KANSTGILSV RDATK ITLA PSSADINLGD NLTLQCHASH DPTMDLTFTW TLDDFPIDFD KPGGHYRRTN VKETIGDLTI LNAQLRHGGK YTCMAQ TVV DSASKEATVL VRGPPGPPGG VVVRDIGDTT IQLSWSRGFD NHSPIAKYTL QARTPPAGKW KQVRTNPANI EGNAETA QV LGLTPWMDYE FRVIASNILG TGEPSGPSSK IRTREAAPSV APSGLSGGGG APGELIVNWT PMSREYQNGD GFGYLLSF R RQGSTHWQTA RVPGADAQYF VYSNESVRPY TPFEVKIRSY NRRGDGPESL TALVYSAEEE PRVAPTKVWA KGVSSSEMN VTWEPVQQDM NGILLGYEIR YWKAGDKEAA ADRVRTAGLD TSARVSGLHP NTKYHVTVRA YNRAGTGPAS PSANATTMKP PPRRPPGNI SWTFSSSSLS IKWDPVVPFR NESAVTGYKM LYQNDLHLTP TLHLTGKNWI EIPVPEDIGH ALVQIRTTGP G GDGIPAEV HIVRNGGTSM MVEN UniProtKB: Contactin-2 |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)