+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of nucleotide-bound ComA with ZinC ion | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC transport / PCAT / MEMBRANE PROTEIN / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationABC-type bacteriocin transporter activity / ABC-type oligopeptide transporter activity / cysteine-type peptidase activity / ATP hydrolysis activity / proteolysis / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Streptococcus pneumoniae D39 (bacteria) Streptococcus pneumoniae D39 (bacteria) | |||||||||
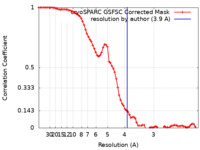
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Yu L / Xin X / Min L / Feng H | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis of peptide secretion for Quorum sensing by ComA. Authors: Lin Yu / Xin Xu / Wan-Zhen Chua / Hao Feng / Zheng Ser / Kai Shao / Jian Shi / Yumei Wang / Zongli Li / Radoslaw M Sobota / Lok-To Sham / Min Luo /    Abstract: Quorum sensing (QS) is a crucial regulatory mechanism controlling bacterial signalling and holds promise for novel therapies against antimicrobial resistance. In Gram-positive bacteria, such as ...Quorum sensing (QS) is a crucial regulatory mechanism controlling bacterial signalling and holds promise for novel therapies against antimicrobial resistance. In Gram-positive bacteria, such as Streptococcus pneumoniae, ComA is a conserved efflux pump responsible for the maturation and secretion of peptide signals, including the competence-stimulating peptide (CSP), yet its structure and function remain unclear. Here, we functionally characterize ComA as an ABC transporter with high ATP affinity and determined its cryo-EM structures in the presence or absence of CSP or nucleotides. Our findings reveal a network of strong electrostatic interactions unique to ComA at the intracellular gate, a putative binding pocket for two CSP molecules, and negatively charged residues facilitating CSP translocation. Mutations of these residues affect ComA's peptidase activity in-vitro and prevent CSP export in-vivo. We demonstrate that ATP-Mg triggers the outward-facing conformation of ComA for CSP release, rather than ATP alone. Our study provides molecular insights into the QS signal peptide secretion, highlighting potential targets for QS-targeting drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36882.map.gz emd_36882.map.gz | 40.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36882-v30.xml emd-36882-v30.xml emd-36882.xml emd-36882.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36882_fsc.xml emd_36882_fsc.xml | 7.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_36882.png emd_36882.png | 70.7 KB | ||
| Filedesc metadata |  emd-36882.cif.gz emd-36882.cif.gz | 5.8 KB | ||
| Others |  emd_36882_half_map_1.map.gz emd_36882_half_map_1.map.gz emd_36882_half_map_2.map.gz emd_36882_half_map_2.map.gz | 39.9 MB 39.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36882 http://ftp.pdbj.org/pub/emdb/structures/EMD-36882 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36882 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36882 | HTTPS FTP |
-Validation report
| Summary document |  emd_36882_validation.pdf.gz emd_36882_validation.pdf.gz | 932.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36882_full_validation.pdf.gz emd_36882_full_validation.pdf.gz | 931.8 KB | Display | |
| Data in XML |  emd_36882_validation.xml.gz emd_36882_validation.xml.gz | 15 KB | Display | |
| Data in CIF |  emd_36882_validation.cif.gz emd_36882_validation.cif.gz | 19.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36882 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36882 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36882 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36882 | HTTPS FTP |
-Related structure data
| Related structure data |  8k4bMC  8hf4C  8hf5C  8hf6C  8hf7C  8k7aC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36882.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36882.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.105 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36882_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_36882_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ComA
| Entire | Name: ComA |
|---|---|
| Components |
|
-Supramolecule #1: ComA
| Supramolecule | Name: ComA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Competence factor transporting ATP-binding protein/permease ComA
| Macromolecule | Name: Competence factor transporting ATP-binding protein/permease ComA type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptococcus pneumoniae D39 (bacteria) / Strain: D39 Streptococcus pneumoniae D39 (bacteria) / Strain: D39 |
| Molecular weight | Theoretical: 80.433031 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKFGKRHYRP QVDQMDCGVA SLAMVFGYYG SYYFLAHLRE LAKTTMDGTT ALGLVKVAEE IGFETRAIKA DMTLFDLPDL TFPFVAHVL KEGKLLHYYV VTGQDKDSIH IADPDPGVKL TKLPRERFEE EWTGVTLFMA PSPDYKPHKE QKNGLLSFIP I LVKQRGLI ...String: MKFGKRHYRP QVDQMDCGVA SLAMVFGYYG SYYFLAHLRE LAKTTMDGTT ALGLVKVAEE IGFETRAIKA DMTLFDLPDL TFPFVAHVL KEGKLLHYYV VTGQDKDSIH IADPDPGVKL TKLPRERFEE EWTGVTLFMA PSPDYKPHKE QKNGLLSFIP I LVKQRGLI ANIVLATLLV TVINIVGSYY LQSIIDTYVP DQMRSTLGII SIGLVIVYIL QQILSYAQEY LLLVLGQRLS ID VILSYIK HVFHLPMSFF ATRRTGEIVS RFTDANSIID ALASTILSIF LDVSTVVIIS LVLFSQNTNL FFMTLLALPI YTV IIFAFM KPFEKMNRDT MEANAVLSSS IIEDINGIET IKSLTSESQR YQKIDKEFVD YLKKSFTYSR AESQQKALKK VAHL LLNVG ILWMGAVLVM DGKMSLGQLI TYNTLLVYFT NPLENIINLQ TKLQTAQVAN NRLNEVYLVA SEFEEKKTVE DLSLM KGDM TFKQVHYKYG YGRDVLSDIN LTVPQGSKVA FVGISGSGKT TLAKMMVNFY DPSQGEISLG GVNLNQIDKK ALRQYI NYL PQQPYVFNGT ILENLLLGAK EGTTQEDILR AVELAEIRED IERMPLNYQT ELTSDGAGIS GGQRQRIALA RALLTDA PV IILDEATSSL DILTEKRIVD NLIALDKTLI FIAHRLTIAE RTEKVVVLDQ GKIVEEGKHA DLLAQGGFYA HLVNS UniProtKB: Transport/processing ATP-binding protein ComA |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 25 mM Tris, pH 7.5, 150 mM NaCl, 2 mM DTT |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)