+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of PseP with NAD at 2.86 angstrom resolution | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | altemicidin / pyridoxal 5'-phosphate / PLP-dependent enzyme / LYASE | |||||||||
| Biological species |  Pseudomonas fluorescens (bacteria) Pseudomonas fluorescens (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.86 Å | |||||||||
 Authors Authors | Mori T / Awakawa T / Adachi N / Abe I | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of PseP with NAD at 2.86 angstrom resolution Authors: Mori T / Awakawa T / Adachi N / Abe I | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36851.map.gz emd_36851.map.gz | 96.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36851-v30.xml emd-36851-v30.xml emd-36851.xml emd-36851.xml | 20.3 KB 20.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36851_fsc.xml emd_36851_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_36851.png emd_36851.png | 44.8 KB | ||
| Masks |  emd_36851_msk_1.map emd_36851_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36851.cif.gz emd-36851.cif.gz | 6.8 KB | ||
| Others |  emd_36851_half_map_1.map.gz emd_36851_half_map_1.map.gz emd_36851_half_map_2.map.gz emd_36851_half_map_2.map.gz | 79.6 MB 79.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36851 http://ftp.pdbj.org/pub/emdb/structures/EMD-36851 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36851 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36851 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36851.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36851.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
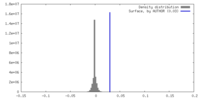
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36851_msk_1.map emd_36851_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
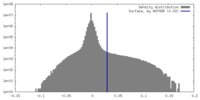
| Density Histograms |
-Half map: #1
| File | emd_36851_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
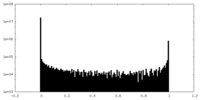
| Density Histograms |
-Half map: #2
| File | emd_36851_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
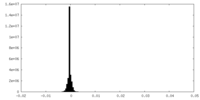
| Density Histograms |
- Sample components
Sample components
-Entire : PseP
| Entire | Name: PseP |
|---|---|
| Components |
|
-Supramolecule #1: PseP
| Supramolecule | Name: PseP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: PLP-dependent enzyme in the biosynthesis of altemicidin |
|---|---|
| Source (natural) | Organism:  Pseudomonas fluorescens (bacteria) Pseudomonas fluorescens (bacteria) |
| Molecular weight | Theoretical: 90 KDa |
-Macromolecule #1: PseP
| Macromolecule | Name: PseP / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas fluorescens (bacteria) Pseudomonas fluorescens (bacteria) |
| Molecular weight | Theoretical: 90.951523 KDa |
| Recombinant expression | Organism:  Streptomyces lividans TK24 (bacteria) Streptomyces lividans TK24 (bacteria) |
| Sequence | String: VNGTFVESIA DAHVELRAAM LGSRHHPSPD IPIINHPSAS GLDQAFALVE ASLTAHGLAI VQMDEPLSTE QFACYARRLG VLVPEHDED VQPFVEQGDI LHLRTRFGPT DRVGLQPFSS SPLSMHSESS GNALVDQPRY LAFQCLEPGE FAYAPQTLLI D MASIVARI ...String: VNGTFVESIA DAHVELRAAM LGSRHHPSPD IPIINHPSAS GLDQAFALVE ASLTAHGLAI VQMDEPLSTE QFACYARRLG VLVPEHDED VQPFVEQGDI LHLRTRFGPT DRVGLQPFSS SPLSMHSESS GNALVDQPRY LAFQCLEPGE FAYAPQTLLI D MASIVARI SPYNINILAR TYYDSQRNSP PLLRYDGQRW VISFRDFQQQ PLSWVHEGPT PAGDVLSAIR DLLACMYTAQ AS AVRWARG MFMVFDNQRY MHARSKGHFV LDQQDRHLLR ARIRARTPDL NVLAAVDDGD SRVLFARPAS GRIPQLPDDF RQT SAVEPN QVEETPDTFI DERTLEVFSR ALNPTNPMEL RNLWLGRVEA ELGDNALRPE YADLWRRSRV RRAVSVEEVL RSTA TVGMV KELFNAFFRD DLYGALSSKR NIILSSGAVD EDEYGLPAAL KETLRFALAR NFYGYSDSLG RQPAREAVAA MESVS MQQG HYEAASVALT MGATHTISSL ADFIFRDNPY ADAAICAIPN YPPLVQSIAW RHPVLLVPTP SHGGTTSLQA LSRAVT PNT PMVLLQTGTN PCGSLVDELE LERFIQSTSL STLIILDECH EWLGAPRHFS PARQRANVIR VSSLS(LLP)NWSV PGL KVGWFL ADPALVSRYY EFASTSYGGP QSFVYTLVEV LARFERWIIE GRTSIDQQQL REFSASYGLQ LGSLSQTYEH YVAE RRARE QVLLGLRGEA TSCLRRASMI VKTPQCSINV FAQIPGSEDS YLSFRNVLRE TGVSVYPGIL SFYLAGGGFR VTTAR KWGD LHRGLERLSA GAGNA |
-Macromolecule #2: NICOTINAMIDE-ADENINE-DINUCLEOTIDE
| Macromolecule | Name: NICOTINAMIDE-ADENINE-DINUCLEOTIDE / type: ligand / ID: 2 / Number of copies: 2 / Formula: NAD |
|---|---|
| Molecular weight | Theoretical: 663.425 Da |
| Chemical component information |  ChemComp-NAD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV |
| Details | This sample was mono-disperse. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 3006 / Average electron dose: 49.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 105000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)