+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CXCR3-DNGi complex activated by CXCL10 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | G protein coupled receptor / chemokine receptor / CXCR3 / CXCL10 / complex / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of T cell chemotaxis / CXCR3 chemokine receptor binding / Oplophorus-luciferin 2-monooxygenase / Oplophorus-luciferin 2-monooxygenase activity / negative regulation of myoblast fusion / regulation of leukocyte migration / chemokine binding / cAMP-dependent protein kinase regulator activity / C-X-C chemokine binding / regulation of endothelial tube morphogenesis ...regulation of T cell chemotaxis / CXCR3 chemokine receptor binding / Oplophorus-luciferin 2-monooxygenase / Oplophorus-luciferin 2-monooxygenase activity / negative regulation of myoblast fusion / regulation of leukocyte migration / chemokine binding / cAMP-dependent protein kinase regulator activity / C-X-C chemokine binding / regulation of endothelial tube morphogenesis / chemokine receptor activity / cellular response to interleukin-17 / CXCR chemokine receptor binding / C-X-C chemokine receptor activity / response to auditory stimulus / positive regulation of chemotaxis / negative regulation of myoblast differentiation / C-C chemokine receptor activity / T cell chemotaxis / chemokine-mediated signaling pathway / C-C chemokine binding / positive regulation of monocyte chemotaxis / negative regulation of execution phase of apoptosis / chemokine activity / response to vitamin D / endothelial cell activation / Chemokine receptors bind chemokines / blood circulation / muscle organ development / chemoattractant activity / negative regulation of endothelial cell proliferation / Interleukin-10 signaling / positive regulation of execution phase of apoptosis / positive regulation of T cell migration / regulation of cell adhesion / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / D2 dopamine receptor binding / response to prostaglandin E / neutrophil chemotaxis / adenylate cyclase regulator activity / G protein-coupled serotonin receptor binding / adenylate cyclase-inhibiting serotonin receptor signaling pathway / antiviral innate immune response / cellular response to forskolin / negative regulation of angiogenesis / regulation of mitotic spindle organization / positive regulation of release of sequestered calcium ion into cytosol / bioluminescence / response to gamma radiation / cell chemotaxis / Regulation of insulin secretion / calcium-mediated signaling / positive regulation of cholesterol biosynthetic process / electron transport chain / negative regulation of insulin secretion / G protein-coupled receptor binding / response to peptide hormone / cellular response to virus / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / chemotaxis / positive regulation of angiogenesis / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / GDP binding / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / cell-cell signaling / GPER1 signaling / G-protein beta-subunit binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Oplophorus gracilirostris (crustacean) Oplophorus gracilirostris (crustacean) | |||||||||
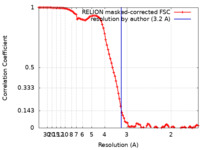
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Jiao HZ / Hu HL | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2023 Journal: Cell Discov / Year: 2023Title: Structure basis for the modulation of CXC chemokine receptor 3 by antagonist AMG487. Authors: Haizhan Jiao / Bin Pang / Ying-Chih Chiang / Qiang Chen / Qi Pan / Ruobing Ren / Hongli Hu /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36842.map.gz emd_36842.map.gz | 66.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36842-v30.xml emd-36842-v30.xml emd-36842.xml emd-36842.xml | 29.1 KB 29.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36842_fsc.xml emd_36842_fsc.xml | 10 KB | Display |  FSC data file FSC data file |
| Images |  emd_36842.png emd_36842.png | 105.6 KB | ||
| Filedesc metadata |  emd-36842.cif.gz emd-36842.cif.gz | 8 KB | ||
| Others |  emd_36842_additional_1.map.gz emd_36842_additional_1.map.gz emd_36842_additional_2.map.gz emd_36842_additional_2.map.gz emd_36842_half_map_1.map.gz emd_36842_half_map_1.map.gz emd_36842_half_map_2.map.gz emd_36842_half_map_2.map.gz | 41.4 MB 65.1 MB 65.4 MB 65.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36842 http://ftp.pdbj.org/pub/emdb/structures/EMD-36842 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36842 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36842 | HTTPS FTP |
-Validation report
| Summary document |  emd_36842_validation.pdf.gz emd_36842_validation.pdf.gz | 813.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36842_full_validation.pdf.gz emd_36842_full_validation.pdf.gz | 813 KB | Display | |
| Data in XML |  emd_36842_validation.xml.gz emd_36842_validation.xml.gz | 17.8 KB | Display | |
| Data in CIF |  emd_36842_validation.cif.gz emd_36842_validation.cif.gz | 22.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36842 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36842 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36842 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36842 | HTTPS FTP |
-Related structure data
| Related structure data |  8k2xMC  8k2wC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36842.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36842.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: local scale map
| File | emd_36842_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local scale map | ||||||||||||
| Projections & Slices |
| ||||||||||||
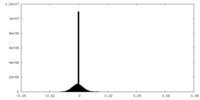
| Density Histograms |
-Additional map: unsharpen map
| File | emd_36842_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpen map | ||||||||||||
| Projections & Slices |
| ||||||||||||
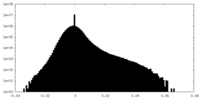
| Density Histograms |
-Half map: #2
| File | emd_36842_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
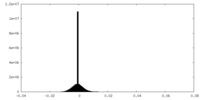
| Density Histograms |
-Half map: #1
| File | emd_36842_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
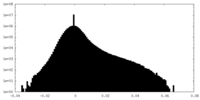
| Density Histograms |
- Sample components
Sample components
-Entire : CXCR3-CXCL10-DNGi-scFv16
| Entire | Name: CXCR3-CXCL10-DNGi-scFv16 |
|---|---|
| Components |
|
-Supramolecule #1: CXCR3-CXCL10-DNGi-scFv16
| Supramolecule | Name: CXCR3-CXCL10-DNGi-scFv16 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.445059 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 42.005895 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHHHHH LEVLFQGPGS SGSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSR LLVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE L AGHTGYLS ...String: HHHHHHHHHH LEVLFQGPGS SGSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSR LLVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE L AGHTGYLS CCRFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CR QTFTGHE SDINAICFFP NGNAFATGSD DATCRLFDLR ADQELMTYSH DNIICGITSV SFSKSGRLLL AGYDDFNCNV WDA LKADRA GVLAGHDNRV SCLGVTDDGM AVATGSWDSF LKIWNGASGA SGASGASVSG WRLFKKIS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Soluble cytochrome b562,C-X-C chemokine receptor type 3,Oplophoru...
| Macromolecule | Name: Soluble cytochrome b562,C-X-C chemokine receptor type 3,Oplophorus-luciferin 2-monooxygenase catalytic subunit type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: Oplophorus-luciferin 2-monooxygenase |
|---|---|
| Source (natural) | Organism:  Oplophorus gracilirostris (crustacean) Oplophorus gracilirostris (crustacean) |
| Molecular weight | Theoretical: 72.918008 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKGSADLE DNWETLNDNL KVIEKADNAA QVKDALTKMR AAALDAQKAT PPKLEDKSPD SPEMKDFRH GFDILVGQID DALKLANEGK VKEAQAAAEQ LKTTRNAYIQ KYLLVPRGSM VLEVSDHQVL NDAEVAALLE N FSSSYDYG ...String: MKTIIALSYI FCLVFADYKD DDDKGSADLE DNWETLNDNL KVIEKADNAA QVKDALTKMR AAALDAQKAT PPKLEDKSPD SPEMKDFRH GFDILVGQID DALKLANEGK VKEAQAAAEQ LKTTRNAYIQ KYLLVPRGSM VLEVSDHQVL NDAEVAALLE N FSSSYDYG ENESDSCCTS PPCPQDFSLN FDRAFLPALY SLLFLLGLLG NGAVAAVLLS RRTALSSTDT FLLHLAVADT LL VLTLPLW AVDAAVQWVF GSGLCKVAGA LFNINFYAGA LLLACISFDR YLNIVHATQL YRRGPPARVT LTCLAVWGLC LLF ALPDFI FLSAHHDERL NATHCQYNFP QVGRTALRVL QLVAGFLLPL LVMAYCYAHI LAVLLVSRGQ RRLRAMRLVV VVVV AFALC WTPYHLVVLV DILMDLGALA RNCGRESRVD VAKSVTSGLG YMHCCLNPLL YAFVGVKFRE RMWMLLLRLG CPNQR GLQR QPSSSRRDSS WSETSVFTLE DFVGDWEQTA AYNLDQVLEQ GGVSSLLQNL AVSVTPIQRI VRSGENALKI DIHVII PYE GLSADQMAQI EEVFKVVYPV DDHHFKVILP YGTLVIDGVT PNMLNYFGRP YEGIAVFDGK KITVTGTLWN GNKIIDE RL ITPDGSMLFR VTINS UniProtKB: Soluble cytochrome b562, C-X-C chemokine receptor type 3, Oplophorus-luciferin 2-monooxygenase catalytic subunit |
-Macromolecule #5: single-chain variable fragment scFv16
| Macromolecule | Name: single-chain variable fragment scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Oplophorus gracilirostris (crustacean) Oplophorus gracilirostris (crustacean) |
| Molecular weight | Theoretical: 27.784896 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAAHHHHHH HH |
-Macromolecule #6: C-X-C motif chemokine 10
| Macromolecule | Name: C-X-C motif chemokine 10 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 10.898089 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNQTAILICC LIFLTLSGIQ GVPLSRTVRC TCISISNQPV NPRSLEKLEI IPASQFCPRV EIIATMKKKG EKRCLNPESK AIKNLLKAV SKERSKRSP UniProtKB: C-X-C motif chemokine 10 |
-Macromolecule #7: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 7 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 53.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)