+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | mycobacterial efflux pump, AMPPNP bound state | |||||||||
 Map data Map data | EM map of mycobacterial efflux pump, AMPPNP bound state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC transporter / efflux pump / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationTranslocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate / response to antibiotic / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Wang Y / Wu F / Zhang L / Rao Z | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Cryo-EM structures of a mycobacterial ABC transporter that mediates rifampicin resistance. Authors: Yinan Wang / Shan Gao / Fangyu Wu / Yicheng Gong / Nengjiang Mu / Chuancun Wei / Chengyao Wu / Jun Wang / Ning Yan / Huifang Yang / Yifan Zhang / Jiayi Liu / Zeyu Wang / Xiuna Yang / Sin Man ...Authors: Yinan Wang / Shan Gao / Fangyu Wu / Yicheng Gong / Nengjiang Mu / Chuancun Wei / Chengyao Wu / Jun Wang / Ning Yan / Huifang Yang / Yifan Zhang / Jiayi Liu / Zeyu Wang / Xiuna Yang / Sin Man Lam / Guanghou Shui / Siyuan Li / Lintai Da / Luke W Guddat / Zihe Rao / Lu Zhang /   Abstract: Drug-resistant Tuberculosis (TB) is a global public health problem. Resistance to rifampicin, the most effective drug for TB treatment, is a major growing concern. The etiological agent, (), has a ...Drug-resistant Tuberculosis (TB) is a global public health problem. Resistance to rifampicin, the most effective drug for TB treatment, is a major growing concern. The etiological agent, (), has a cluster of ATP-binding cassette (ABC) transporters which are responsible for drug resistance through active export. Here, we describe studies characterizing Rv1217c-1218c as an ABC transporter that can mediate mycobacterial resistance to rifampicin and have determined the cryo-electron microscopy structures of Rv1217c-1218c. The structures show Rv1217c-1218c has a type V exporter fold. In the absence of ATP, Rv1217c-1218c forms a periplasmic gate by two juxtaposed-membrane helices from each transmembrane domain (TMD), while the nucleotide-binding domains (NBDs) form a partially closed dimer which is held together by four salt-bridges. Adenylyl-imidodiphosphate (AMPPNP) binding induces a structural change where the NBDs become further closed to each other, which downstream translates to a closed conformation for the TMDs. AMPPNP binding results in the collapse of the outer leaflet cavity and the opening of the periplasmic gate, which was proposed to play a role in substrate export. The rifampicin-bound structure shows a hydrophobic and periplasm-facing cavity is involved in rifampicin binding. Phospholipid molecules are observed in all determined structures and form an integral part of the Rv1217c-1218c transporter system. Our results provide a structural basis for a mycobacterial ABC exporter that mediates rifampicin resistance, which can lead to different insights into combating rifampicin resistance. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36797.map.gz emd_36797.map.gz | 203.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36797-v30.xml emd-36797-v30.xml emd-36797.xml emd-36797.xml | 22.5 KB 22.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36797.png emd_36797.png | 45.8 KB | ||
| Filedesc metadata |  emd-36797.cif.gz emd-36797.cif.gz | 7.2 KB | ||
| Others |  emd_36797_half_map_1.map.gz emd_36797_half_map_1.map.gz emd_36797_half_map_2.map.gz emd_36797_half_map_2.map.gz | 200.6 MB 200.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36797 http://ftp.pdbj.org/pub/emdb/structures/EMD-36797 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36797 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36797 | HTTPS FTP |
-Validation report
| Summary document |  emd_36797_validation.pdf.gz emd_36797_validation.pdf.gz | 809.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36797_full_validation.pdf.gz emd_36797_full_validation.pdf.gz | 809.5 KB | Display | |
| Data in XML |  emd_36797_validation.xml.gz emd_36797_validation.xml.gz | 15.5 KB | Display | |
| Data in CIF |  emd_36797_validation.cif.gz emd_36797_validation.cif.gz | 18.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36797 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36797 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36797 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36797 | HTTPS FTP |
-Related structure data
| Related structure data |  8k1oMC  8k1mC  8k1nC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36797.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36797.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map of mycobacterial efflux pump, AMPPNP bound state | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
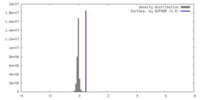
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map B of mycobacterial efflux pump, AMPPNP bound state
| File | emd_36797_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B of mycobacterial efflux pump, AMPPNP bound state | ||||||||||||
| Projections & Slices |
| ||||||||||||
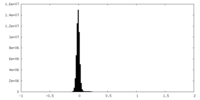
| Density Histograms |
-Half map: half map A of mycobacterial efflux pump, AMPPNP bound state
| File | emd_36797_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A of mycobacterial efflux pump, AMPPNP bound state | ||||||||||||
| Projections & Slices |
| ||||||||||||
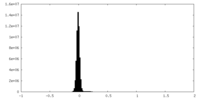
| Density Histograms |
- Sample components
Sample components
-Entire : ternary complex of an ABC transporter Rv1217c-1218c
| Entire | Name: ternary complex of an ABC transporter Rv1217c-1218c |
|---|---|
| Components |
|
-Supramolecule #1: ternary complex of an ABC transporter Rv1217c-1218c
| Supramolecule | Name: ternary complex of an ABC transporter Rv1217c-1218c / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) |
| Molecular weight | Theoretical: 120 KDa |
-Macromolecule #1: Multidrug efflux system permease protein Rv1217c
| Macromolecule | Name: Multidrug efflux system permease protein Rv1217c / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) |
| Molecular weight | Theoretical: 56.698039 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MSSTVIDRAR PAGHRAPHRG SGFTGTLGLL RLYLRRDRVS LPLWVLLLSV PLATVYIASV ETVYPDRSAR AAAAAAIMAS PAQRALYGP VYNDSLGAVG IWKAGMFHTL IAVAVILTVI RHTRADEESG RAELIDSTVV GRYANLTGAL LLSFGASIAT G AIGALGLL ...String: MSSTVIDRAR PAGHRAPHRG SGFTGTLGLL RLYLRRDRVS LPLWVLLLSV PLATVYIASV ETVYPDRSAR AAAAAAIMAS PAQRALYGP VYNDSLGAVG IWKAGMFHTL IAVAVILTVI RHTRADEESG RAELIDSTVV GRYANLTGAL LLSFGASIAT G AIGALGLL ATDVAPAGSV AFGVALAASG MVFTAVAAVA AQLSPSARFT RAVAFAVLGT AFALRAIGDA GSGTLSWCSP LG WSLQVRP YAGERWWVLL LSLATAAVLT VLAYRLRAGR DVGAGLIAER PGAGTAGPML SEPFGLAWRL NRGSLLLWTV GLC LYGLVM GSVVHGIGDQ LGDNTAVRDI VTRMGGTGAL EQAFLALAFT MIGMVAAAFA VSLTLRLHQE ETGLRAETLL AGAV SRTHW LASHLAMALA GSAVATLISG VAAGLAYGMT VGDVGGKLPT VVGTAAVQLP AVWLLSAVTV GLFGLAPRFT PVAWG VLVG FIALYLLGSL AGFPQMLLNL EPFAHIPRVG GGDFTAVPLL WLLAIDAALI TLGAMAFRRR DVRC UniProtKB: Multidrug efflux system permease protein Rv1217c |
-Macromolecule #2: Multidrug efflux system ATP-binding protein Rv1218c
| Macromolecule | Name: Multidrug efflux system ATP-binding protein Rv1218c / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO EC number: Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) |
| Molecular weight | Theoretical: 33.491082 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MSADNHQVPI EIRGLTKHFG SVRALDGLDL TVREGEVHGF LGPNGAGKST TLRILLGLVK ADGGSVRLLG GDPWTDAVDL HRHIAYVPG DVTLWPSLTG GETIDLLARM RGGIDNARRA ELIERFGLDP TKKARTYSKG NRQKVSLISA LSSHATLLLL D EPSSGLDP ...String: MSADNHQVPI EIRGLTKHFG SVRALDGLDL TVREGEVHGF LGPNGAGKST TLRILLGLVK ADGGSVRLLG GDPWTDAVDL HRHIAYVPG DVTLWPSLTG GETIDLLARM RGGIDNARRA ELIERFGLDP TKKARTYSKG NRQKVSLISA LSSHATLLLL D EPSSGLDP LMENVFQQCI GEARQRGVTV LLSSHILAET EALCEKVTII RAGKTVESGS LDALRHLSRT SIKAEMIGDP GD LSQIKGV EDISIEGTTV RAQVDSESLR ELIQVLGHAG VRSLVSQPPT LEELFLRHYS LGPEVAAEQQ VATP UniProtKB: Multidrug efflux system ATP-binding protein Rv1218c |
-Macromolecule #3: CARDIOLIPIN
| Macromolecule | Name: CARDIOLIPIN / type: ligand / ID: 3 / Number of copies: 2 / Formula: CDL |
|---|---|
| Molecular weight | Theoretical: 1.464043 KDa |
| Chemical component information |  ChemComp-CDL: |
-Macromolecule #4: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 4 / Number of copies: 2 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 150mM NaCl, 20mM Tris, detergent |
| Grid | Material: COPPER / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK II |
| Details | sample mono disperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 3629 / Average exposure time: 2.4 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)