[English] 日本語
 Yorodumi
Yorodumi- EMDB-36794: Cryo-EM structure of Na+,K+-ATPase alpha2 from Artemia salina in ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Na+,K+-ATPase alpha2 from Artemia salina in cation-free E2P form | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | P-type ATPase / sodium pump / membrane protein / transporter / TRANSPORT PROTEIN | |||||||||
| Biological species |  Artemia salina (crustacean) Artemia salina (crustacean) | |||||||||
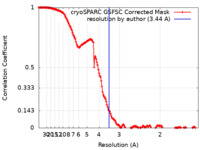
| Method | single particle reconstruction / cryo EM / Resolution: 3.44 Å | |||||||||
 Authors Authors | Abe K / Artigas P | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: A Na pump with reduced stoichiometry is up-regulated by brine shrimp in extreme salinities. Authors: Pablo Artigas / Dylan J Meyer / Victoria C Young / Kerri Spontarelli / Jessica Eastman / Evan Strandquist / Huan Rui / Benoît Roux / Matthew A Birk / Hanayo Nakanishi / Kazuhiro Abe / Craig Gatto /   Abstract: Brine shrimp () are the only animals to thrive at sodium concentrations above 4 M. Salt excretion is powered by the Na,K-ATPase (NKA), a heterodimeric (αβ) pump that usually exports 3Na in exchange ...Brine shrimp () are the only animals to thrive at sodium concentrations above 4 M. Salt excretion is powered by the Na,K-ATPase (NKA), a heterodimeric (αβ) pump that usually exports 3Na in exchange for 2 K per hydrolyzed ATP. express several NKA catalytic α-subunit subtypes. High-salinity adaptation increases abundance of α2, an isoform that contains two lysines (Lys308 and Lys758 in transmembrane segments TM4 and TM5, respectively) at positions where canonical NKAs have asparagines ( α1's Asn333 and Asn785). Using de novo transcriptome assembly and qPCR, we found that express two salinity-independent canonical α subunits (α1 and α3), as well as two β variants, in addition to the salinity-controlled α2. These β subunits permitted heterologous expression of the α2 pump and determination of its CryoEM structure in a closed, ion-free conformation, showing Lys758 residing within the ion-binding cavity. We used electrophysiology to characterize the function of α2 pumps and compared it to that of α1 (and its α2-mimicking single- and double-lysine substitutions). The double substitution N333K/N785K confers α2-like characteristics to α1, and mutant cycle analysis reveals energetic coupling between these two residues, illustrating how α2's Lys308 helps to maintain high affinity for external K when Lys758 occupies an ion-binding site. By measuring uptake under voltage clamp of the K-congener Rb, we prove that double-lysine-substituted pumps transport 2Na and 1 K per catalytic cycle. Our results show how the two lysines contribute to generate a pump with reduced stoichiometry allowing to maintain steeper Na gradients in hypersaline environments. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36794.map.gz emd_36794.map.gz | 328.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36794-v30.xml emd-36794-v30.xml emd-36794.xml emd-36794.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36794_fsc.xml emd_36794_fsc.xml | 14.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_36794.png emd_36794.png | 62.4 KB | ||
| Masks |  emd_36794_msk_1.map emd_36794_msk_1.map | 347.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36794.cif.gz emd-36794.cif.gz | 6.2 KB | ||
| Others |  emd_36794_half_map_1.map.gz emd_36794_half_map_1.map.gz emd_36794_half_map_2.map.gz emd_36794_half_map_2.map.gz | 322.3 MB 322.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36794 http://ftp.pdbj.org/pub/emdb/structures/EMD-36794 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36794 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36794 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36794.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36794.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.752 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36794_msk_1.map emd_36794_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_36794_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_36794_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Na+,K+-ATPase alpha2/beta2 from Artemia Salina
| Entire | Name: Na+,K+-ATPase alpha2/beta2 from Artemia Salina |
|---|---|
| Components |
|
-Supramolecule #1: Na+,K+-ATPase alpha2/beta2 from Artemia Salina
| Supramolecule | Name: Na+,K+-ATPase alpha2/beta2 from Artemia Salina / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Artemia salina (crustacean) Artemia salina (crustacean) |
| Molecular weight | Theoretical: 130 KDa |
-Macromolecule #1: Na+,K+-ATPase alpha2KK
| Macromolecule | Name: Na+,K+-ATPase alpha2KK / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Artemia salina (crustacean) Artemia salina (crustacean) |
| Molecular weight | Theoretical: 111.144211 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGKKQGKQLS DLKKELELDQ HKIPLEELCR RLGTNTETGL TSSQAKSHLE KYGPNALTPP RTTPEWIKFC KQLFGGFQML LWIGSILCF IAYTMEKYKN PDVLGDNLYL GLALLFVVIM TGCFAYYQDH NASKIMDSFK NLMPQFAFVI RDGKKIQLKA E EVTVGDLV ...String: MGKKQGKQLS DLKKELELDQ HKIPLEELCR RLGTNTETGL TSSQAKSHLE KYGPNALTPP RTTPEWIKFC KQLFGGFQML LWIGSILCF IAYTMEKYKN PDVLGDNLYL GLALLFVVIM TGCFAYYQDH NASKIMDSFK NLMPQFAFVI RDGKKIQLKA E EVTVGDLV EVKFGDRIPA DIRITSCQSM KVDNSSLTGE SEPQSRSTEC TNDNPLETKN LAFFFTNTLE GTGRGIVINV GD DSVMGRI ACLASSLDSG KTPIAREIEH FIHIITAMAV SLAAVFAVIS FLYGYTWLEA AIFMIGIIVA KVPEGLLATV TVC LTLTAK RMAKKNCLVR NLEAVETLGS TSTICSDKTG TLTQNRMTVA HMWFDQKIVT ADTTENQSGN QLYRGSKGFP ELIR VASLC SRAEFKTEHA HLPVLKRDVN GDASEAAILK FAEMSTGSVM NIRSKQKKVS EIPFNSANKY QVSVHEREDK SGYFL VMKG APERILERCS TILIDGTEIP LDNHMKECFN NAYMELGGMG ERVLGFCDFE LPSDQYPRGY VFDADEPNFP ISGLRF VGL MSMIDPPRAA VPDAVSKCRS AGIKVIMVTG DHPITAKAIA RQVGIISEGH ETVDDIAARL NIPVSEVNPR SAQAAVI HG NDLKDMNSDQ LDDILRHYRE IVFARTSPQQ KLIIVEGVQR QGEFVAVTGD GVNDSPALKK ADIGVAMGIA GSDVSKQA A DMILLDDNFA SIVTGVEEGR LIFDNIKKSI AYTLTSKIPE LSPFLMYILF DLPLAIGTVT ILCIDLGTDV VPAISMAYE GPEADPRKPR DPVKEKLVNE RLISMAYGQI GVMQAFGGFF TYFVIMGECG FLPNRLFGLR KWWESKAYND LTDSYGQEWT WDARKQLEY TCHTAFFISI VIVQWTDLII CKTRRLSLFQ QGMKNGTLNF ALVFETCVAA FLSYTPGMDK GLRMYPLKIW W WFPPMPFS LLILVYDECR KFLMRRNPGG FLERETYY |
-Macromolecule #2: Na+,K+-ATPase beta2
| Macromolecule | Name: Na+,K+-ATPase beta2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Artemia salina (crustacean) Artemia salina (crustacean) |
| Molecular weight | Theoretical: 38.496422 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGDYKDDDDK SSGENLYFQG MADKKPEEFF VGSGPKPTKW QSVKTFIWNS ETSEFMGRTG VNWAKITIFY VIFYTLLAGF FAGMLMIFY QTLDFKIPKW QNKDSLIGTN PGLGFRPMPP EAQVDSTLIQ FKHGIKGDWQ YWVHSLTEFL EPYETLTSSG Q EFTNCDFD ...String: MGDYKDDDDK SSGENLYFQG MADKKPEEFF VGSGPKPTKW QSVKTFIWNS ETSEFMGRTG VNWAKITIFY VIFYTLLAGF FAGMLMIFY QTLDFKIPKW QNKDSLIGTN PGLGFRPMPP EAQVDSTLIQ FKHGIKGDWQ YWVHSLTEFL EPYETLTSSG Q EFTNCDFD KPPQEGKACN FNVELLGDHC TKENNFGYEL GKPCVLIKLN KIFGWRPEVY NSSAEVPEDM PADLKSYIKD IE TGNKTHM NMVWLSCEGE TANDKEKIGT ITYTPFRGFP AYYYPYLNVP GYLTPVVALQ FGSLQNGQAV NVECKAWANN ISR DRQRRL GSVHFEIRMD |
-Macromolecule #3: TETRAFLUOROALUMINATE ION
| Macromolecule | Name: TETRAFLUOROALUMINATE ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: ALF |
|---|---|
| Molecular weight | Theoretical: 102.975 Da |
| Chemical component information |  ChemComp-ALF: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)