[English] 日本語
 Yorodumi
Yorodumi- EMDB-36758: CryoEM structure of 3-phenylpropionate/cinnamic acid dioxygenase ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of 3-phenylpropionate/cinnamic acid dioxygenase HcaE-HcaF complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | 3-phenylpropionate/cinnamic acid dioxygenase / BIOSYNTHETIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information3-phenylpropanoate dioxygenase / 3-phenylpropionate dioxygenase activity / 3-phenylpropionate dioxygenase complex / 3-phenylpropionate catabolic process / 2 iron, 2 sulfur cluster binding / iron ion binding Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.12 Å | |||||||||
 Authors Authors | Jiang WX / Cheng XQ / Ma LX / Xing Q | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: CryoEM structure of 3-phenylpropionate/cinnamic acid dioxygenase HcaE-HcaF complex Authors: Jiang WX / Cheng XQ / Wu M / Ma LX / Xing Q | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36758.map.gz emd_36758.map.gz | 490.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36758-v30.xml emd-36758-v30.xml emd-36758.xml emd-36758.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36758.png emd_36758.png | 32.5 KB | ||
| Filedesc metadata |  emd-36758.cif.gz emd-36758.cif.gz | 5.5 KB | ||
| Others |  emd_36758_half_map_1.map.gz emd_36758_half_map_1.map.gz emd_36758_half_map_2.map.gz emd_36758_half_map_2.map.gz | 928.2 MB 928.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36758 http://ftp.pdbj.org/pub/emdb/structures/EMD-36758 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36758 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36758 | HTTPS FTP |
-Related structure data
| Related structure data |  8k0aMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36758.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36758.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.425 Å | ||||||||||||||||||||||||||||||||||||
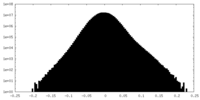
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_36758_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
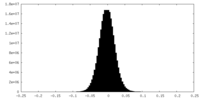
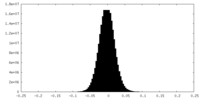
| Density Histograms |
-Half map: #2
| File | emd_36758_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
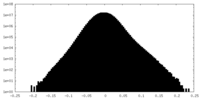
| Density Histograms |
- Sample components
Sample components
-Entire : Homohexamer of inositol phosphate phosphatase SopB
| Entire | Name: Homohexamer of inositol phosphate phosphatase SopB |
|---|---|
| Components |
|
-Supramolecule #1: Homohexamer of inositol phosphate phosphatase SopB
| Supramolecule | Name: Homohexamer of inositol phosphate phosphatase SopB / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: 3-phenylpropionate/cinnamic acid dioxygenase subunit alpha
| Macromolecule | Name: 3-phenylpropionate/cinnamic acid dioxygenase subunit alpha type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO / EC number: 3-phenylpropanoate dioxygenase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 51.166047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTTPSDLNIY QLIDTQNGRV TPRIYTDPDI YQLELERIFG RCWLFLAHES QIPKPGDFFN TYMGEDAVVV VRQKDGSIKA FLNQCRHRA MRVSYADCGN TRAFTCPYHG WSYGINGELI DVPLEPRAYP QGLCKSHWGL NEVPCVESYK GLIFGNWDTS A PGLRDYLG ...String: MTTPSDLNIY QLIDTQNGRV TPRIYTDPDI YQLELERIFG RCWLFLAHES QIPKPGDFFN TYMGEDAVVV VRQKDGSIKA FLNQCRHRA MRVSYADCGN TRAFTCPYHG WSYGINGELI DVPLEPRAYP QGLCKSHWGL NEVPCVESYK GLIFGNWDTS A PGLRDYLG DIAWYLDGML DRREGGTEIV GGVQKWVINC NWKFPAEQFA SDQYHALFSH ASAVQVLGAK DDGSDKRLGD GQ TARPVWE TAKDALQFGQ DGHGSGFFFT EKPDANVWVD GAVSSYYRET YAEAEQRLGE VRALRLAGHN NIFPTLSWLN GTA TLRVWH PRGPDQVEVW AFCITDKAAS DEVKAAFENS ATRAFGPAGF LEQDDSENWC EIQKLLKGHR ARNSKLCLEM GLGQ EKRRD DGIPGITNYI FSETAARGMY QRWADLLSSE SWQEVLDKTA AYQQEVMK UniProtKB: 3-phenylpropionate/cinnamic acid dioxygenase subunit alpha |
-Macromolecule #2: 3-phenylpropionate/cinnamic acid dioxygenase subunit beta
| Macromolecule | Name: 3-phenylpropionate/cinnamic acid dioxygenase subunit beta type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO / EC number: 3-phenylpropanoate dioxygenase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20.609248 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSAQVSLELH HRISQFLFHE ASLLDDWKFR DWLAQLDEEI RYTMRTTVNA QTRDRRKGVQ PPTTWIFNDT KDQLERRIAR LETGMAWAE EPPSRTRHLI SNCQISETDI PNVFAVRVNY LLYRAQKERD ETFYVGTRFD KVRRLEDDNW RLLERDIVLD Q AVITSHNL SVLF UniProtKB: 3-phenylpropionate/cinnamic acid dioxygenase subunit beta |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 39.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.12 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 30478 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)