[English] 日本語
 Yorodumi
Yorodumi- EMDB-36674: Cryo-EM structure of Plasmodium falciparum multidrug resistance p... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Plasmodium falciparum multidrug resistance protein 1 in the apo state with H1 helix | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Plasmodium falciparum / Multidrug resistance protein 1 / ABC transporter / Apo state / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationRecycling of bile acids and salts / Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol / Atorvastatin ADME / Prednisone ADME / ABC-family proteins mediated transport / food vacuole / ABC-type xenobiotic transporter / vacuolar membrane / ATPase-coupled transmembrane transporter activity / ABC-type transporter activity ...Recycling of bile acids and salts / Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol / Atorvastatin ADME / Prednisone ADME / ABC-family proteins mediated transport / food vacuole / ABC-type xenobiotic transporter / vacuolar membrane / ATPase-coupled transmembrane transporter activity / ABC-type transporter activity / transmembrane transport / ATP hydrolysis activity / ATP binding / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.19 Å | |||||||||
 Authors Authors | Li M / Si K | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: The structure of multidrug resistance protein 1 reveals an N-terminal regulatory domain. Authors: Kaixue Si / Xishuo He / Liping Chen / Anqi Zhang / Changyou Guo / Minghui Li /  Abstract: multidrug resistance protein 1 (PfMDR1), an adenosine triphosphate (ATP)-binding cassette (ABC) transporter on the digestive vacuole (DV) membrane of the parasite, is associated with the resistance ... multidrug resistance protein 1 (PfMDR1), an adenosine triphosphate (ATP)-binding cassette (ABC) transporter on the digestive vacuole (DV) membrane of the parasite, is associated with the resistance to antimalarial drugs. To understand the mechanisms of PfMDR1, we determined the cryo-electron microscopy structures of this transporter in different states. The transporter in the apo state shows an inward-facing conformation with a large cavity opening to the cytoplasm. Upon ATP binding and dimerization of the nucleotide-binding domains (NBDs), PfMDR1 displays an outward-facing conformation with a cavity toward the DV lumen. Drug resistance-associated mutations were investigated in both structures for their effects, and Y184F was identified as an allosteric activity-enhancing mutation. The amphiphilic substrate-binding site of PfMDR1 was revealed by the complex structure with the antimalarial drug mefloquine and confirmed by mutagenesis studies. Remarkably, a helical structure was found to hinder NBD dimerization and inhibit PfMDR1 activity. The location of this regulatory domain in the N terminus is different from the well-studied R domain in the internal linker region of other ABC transporter family members. The lack of the phosphorylation site of this domain also suggests a different regulation mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36674.map.gz emd_36674.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36674-v30.xml emd-36674-v30.xml emd-36674.xml emd-36674.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36674.png emd_36674.png | 61.2 KB | ||
| Others |  emd_36674_half_map_1.map.gz emd_36674_half_map_1.map.gz emd_36674_half_map_2.map.gz emd_36674_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36674 http://ftp.pdbj.org/pub/emdb/structures/EMD-36674 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36674 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36674 | HTTPS FTP |
-Related structure data
| Related structure data |  8jvhMC  8jw4C  8jwfC  8jwgC  8jwiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36674.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36674.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_36674_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
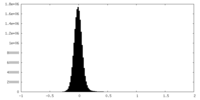
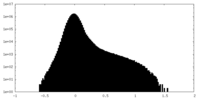
| Density Histograms |
-Half map: #2
| File | emd_36674_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
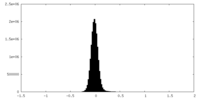
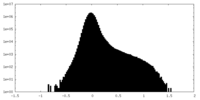
| Density Histograms |
- Sample components
Sample components
-Entire : Plasmodium falciparum multidrug resistance protein 1 in apo state
| Entire | Name: Plasmodium falciparum multidrug resistance protein 1 in apo state |
|---|---|
| Components |
|
-Supramolecule #1: Plasmodium falciparum multidrug resistance protein 1 in apo state
| Supramolecule | Name: Plasmodium falciparum multidrug resistance protein 1 in apo state type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Multidrug resistance protein 1
| Macromolecule | Name: Multidrug resistance protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 162.764812 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GGGGSMGKEQ KEKKDGNLSI KEEVEKELNK KSTAELFRKI KNEKISFFLP FKCLPAQHRK LLFISFVCAV LSGGTLPFFI SVFGVILKN MNLGDDINPI ILSLVSIGLV QFILSMISSY CMDVITSKIL KTLKLEYLRS VFYQDGQFHD NNPGSKLRSD L DFYLEQVS ...String: GGGGSMGKEQ KEKKDGNLSI KEEVEKELNK KSTAELFRKI KNEKISFFLP FKCLPAQHRK LLFISFVCAV LSGGTLPFFI SVFGVILKN MNLGDDINPI ILSLVSIGLV QFILSMISSY CMDVITSKIL KTLKLEYLRS VFYQDGQFHD NNPGSKLRSD L DFYLEQVS SGIGTKFITI FTYASSFLGL YIWSLIKNAR LTLCITCVFP LIYVCGVICN KKVKLNKKTS LLYNNNTMSI IE EALMGIR TVASYCGEKT ILNKFNLSET FYSKYILKAN FVEALHIGLI NGLILVSYAF GFWYGTRIII NSATNQYPNN DFN GASVIS ILLGVLISMF MLTIILPNIT EYMKALEATN SLYEIINRKP LVENNDDGET LPNIKKIEFK NVRFHYDTRK DVEI YKDLS FTLKEGKTYA FVGESGCGKS TILKLIERLY DPTEGDIIVN DSHNLKDINL KWWRSKIGVV SQDPLLFSNS IKNNI KYSL YSLKDLEAME NYYEENTNDT YENKNFSLIS NSMTSNELLE MKKEYQTIKD SDVVDVSKKV LIHDFVSSLP DKYDTL VGS NASKLSGGQK QRISIARAIM RNPKILILDE ATSSLDNKSE YLVQKTINNL KGNENRITII IAHRLSTIRY ANTIFVL SN RERSDNNNNN NNDDNNNNNN NNNNKINNEG SYIIEQGTHD SLMKNKNGIY HLMINNQKIS SNKSSNNGND NGSDNKSS A YKDSDTGNDA DNMNSLSIHE NENISNNRNC KNTAENEKEE KVPFFKRMFR RKKKAPNNLR IIYKEIFSYK KDVTIIFFS ILVAGGLYPV FALLYARYVS TLFDFANLEY NSNKYSIYIL LIAIAMFISE TLKNYYNNKI GEKVEKTMKR RLFENILYQE MSFFDQDKN TPGVLSAHIN RDVHLLKTGL VNNIVIFSHF IMLFLVSMVM SFYFCPIVAA VLTFIYFINM RVFAVRARLT K SKEIEKKE NMSSGVFAFS SDDEMFKDPS FLIQEAFYNM HTVINYGLED YFCNLIEKAI DYKNKGQKRR IIVNAALWGF SQ SAQLFIN SFAYWFGSFL IKRGTILVDD FMKSLFTFIF TGSYAGKLMS LKGDSENAKL SFEKYYPLMI RKSNIDVRDD GGI RINKNL IKGKVDIKDV NFRYISRPNV PIYKNLSFTC DSKKTTAIVG ETGSGKSTFM NLLLRFYDLK NDHIILKNDM TNFQ DYQNN NNNSLVLKNV NEFSNQSGSA EDYTVFNNNG EILLDDINIC DYNLRDLRNL FSIVSQEPML FNMSIYENIK FGRED ATLE DVKRVSKFAA IDEFIESLPN KYDTNVGPYG KSLSGGQKQR IAIARALLRE PKILLLDEAT SSLDSNSEKL IEKTIV DIK DKADKTIITI AHRIASIKRS DKIVVFNNPD RNGTFVQSHG THDELLSAQD GIYKKYVKLA K UniProtKB: Multidrug resistance protein 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.19 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 217767 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)