[English] 日本語
 Yorodumi
Yorodumi- EMDB-36641: BJOX2000.664 trimer in complex with Fab fragment of broadly neutr... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | BJOX2000.664 trimer in complex with Fab fragment of broadly neutralizing HIV antibody PGT145 | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | HIV / Envelope trimer / broadly neutralizing antibody / PGT145 / Cryo-EM / VIRAL PROTEIN | ||||||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
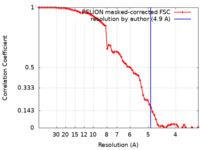
| Method | single particle reconstruction / cryo EM / Resolution: 4.9 Å | ||||||||||||
 Authors Authors | Chatterjee A / Chen C / Lee K / Mangala Prasad V | ||||||||||||
| Funding support |  United States, United States,  India, 3 items India, 3 items
| ||||||||||||
 Citation Citation |  Journal: Npj Viruses / Year: 2023 Journal: Npj Viruses / Year: 2023Title: An HIV-1 broadly neutralizing antibody overcomes structural and dynamic variation through highly focused epitope targeting. Authors: Edgar A Hodge / Ananya Chatterjee / Chengbo Chen / Gajendra S Naika / Mint Laohajaratsang / Vidya Mangala Prasad / Kelly K Lee /   Abstract: The existence of broadly cross-reactive antibodies that can neutralize diverse HIV-1 isolates (bnAbs) has been appreciated for more than a decade. Many high-resolution structures of bnAbs, typically ...The existence of broadly cross-reactive antibodies that can neutralize diverse HIV-1 isolates (bnAbs) has been appreciated for more than a decade. Many high-resolution structures of bnAbs, typically with one or two well-characterized HIV-1 Env glycoprotein trimers, have been reported. However, an understanding of how such antibodies grapple with variability in their antigenic targets across diverse viral isolates has remained elusive. To achieve such an understanding requires first characterizing the extent of structural and antigenic variation embodied in Env, and then identifying how a bnAb overcomes that variation at a structural level. Here, using hydrogen/deuterium-exchange mass spectrometry (HDX-MS) and quantitative measurements of antibody binding kinetics, we show that variation in structural ordering in the V1/V2 apex of Env across a globally representative panel of HIV-1 isolates has a marked effect on antibody association rates and affinities. We also report cryo-EM reconstructions of the apex-targeting PGT145 bnAb bound to two divergent Env that exhibit different degrees of structural dynamics throughout the trimer structures. Parallel HDX-MS experiments demonstrate that PGT145 bnAb has an exquisitely focused footprint at the trimer apex where binding did not yield allosteric changes throughout the rest of the structure. These results demonstrate that structural dynamics are a cryptic determinant of antigenicity, and mature antibodies that have achieved breadth and potency in some cases are able to achieve their broad cross-reactivity by "threading the needle" and binding in a highly focused fashion, thus evading and overcoming the variable properties found in Env from divergent isolates. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36641.map.gz emd_36641.map.gz | 30.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36641-v30.xml emd-36641-v30.xml emd-36641.xml emd-36641.xml | 20.4 KB 20.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36641_fsc.xml emd_36641_fsc.xml | 7.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_36641.png emd_36641.png | 30.2 KB | ||
| Filedesc metadata |  emd-36641.cif.gz emd-36641.cif.gz | 7.1 KB | ||
| Others |  emd_36641_half_map_1.map.gz emd_36641_half_map_1.map.gz emd_36641_half_map_2.map.gz emd_36641_half_map_2.map.gz | 24.8 MB 24.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36641 http://ftp.pdbj.org/pub/emdb/structures/EMD-36641 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36641 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36641 | HTTPS FTP |
-Validation report
| Summary document |  emd_36641_validation.pdf.gz emd_36641_validation.pdf.gz | 660.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36641_full_validation.pdf.gz emd_36641_full_validation.pdf.gz | 660 KB | Display | |
| Data in XML |  emd_36641_validation.xml.gz emd_36641_validation.xml.gz | 12.6 KB | Display | |
| Data in CIF |  emd_36641_validation.cif.gz emd_36641_validation.cif.gz | 17.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36641 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36641 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36641 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36641 | HTTPS FTP |
-Related structure data
| Related structure data |  8jtdMC  8jtmC  36630 M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36641.map.gz / Format: CCP4 / Size: 32.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36641.map.gz / Format: CCP4 / Size: 32.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.72 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_36641_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_36641_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BJOX2000.664 trimer in complex with broadly neutralizing HIV anti...
| Entire | Name: BJOX2000.664 trimer in complex with broadly neutralizing HIV antibody PGT145 |
|---|---|
| Components |
|
-Supramolecule #1: BJOX2000.664 trimer in complex with broadly neutralizing HIV anti...
| Supramolecule | Name: BJOX2000.664 trimer in complex with broadly neutralizing HIV antibody PGT145 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: gp120 protein of HIV Envelope trimer
| Macromolecule | Name: gp120 protein of HIV Envelope trimer / type: protein_or_peptide / ID: 1 Details: The HIV trimer map of this study is of BJOX2000.664 but we have given a sequence of BG505.664 as the model of BG505.664 was used to rigid body fit in our map. Because of the low resolution ...Details: The HIV trimer map of this study is of BJOX2000.664 but we have given a sequence of BG505.664 as the model of BG505.664 was used to rigid body fit in our map. Because of the low resolution of the map, we have not built the model. Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 54.064277 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT ...String: AENLWVTVYY GVPVWKDAET TLFCASDAKA YETEKHNVWA THACVPTDPN PQEIHLENVT EEFNMWKNNM VEQMHTDIIS LWDQSLKPC VKLTPLCVTL QCTNVTNNIT DDMRGELKNC SFNMTTELRD KKQKVYSLFY RLDVVQINEN QGNRSNNSNK E YRLINCNT SAITQACPKV SFEPIPIHYC APAGFAILKC KDKKFNGTGP CPSVSTVQCT HGIKPVVSTQ LLLNGSLAEE EV MIRSENI TNNAKNILVQ FNTPVQINCT RPNNNTRKSI RIGPGQAFYA TGDIIGDIRQ AHCNVSKATW NETLGKVVKQ LRK HFGNNT IIRFANSSGG DLEVTTHSFN CGGEFFYCNT SGLFNSTWIS NTSVQGSNST GSNDSITLPC RIKQIINMWQ RIGQ AMYAP PIQGVIRCVS NITGLILTRD GGSTNSTTET FRPGGGDMRD NWRSELYKYK VVKIEPLGVA PTRCKRRVVG RRRRR R |
-Macromolecule #2: gp41 protein of HIV Envelope trimer
| Macromolecule | Name: gp41 protein of HIV Envelope trimer / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.146482 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEA QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD |
-Macromolecule #3: PGT145 antibody fragment, heavy chain
| Macromolecule | Name: PGT145 antibody fragment, heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.793164 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QASTMDWIWR ILFLVAAATS AHSQVQLVQS GAEVKKPGSS VKVSCKASGN SFSNHDVHWV RQATGQGLEW MGWMSHEGDK TGLAQKFQG RVTITRDSGA STVYMELRGL TADDTAIYYC LTGSKHRLRD YFLYNEYGPN YEEWGDYLAT LDVWGHGTAV T VSSASTKG ...String: QASTMDWIWR ILFLVAAATS AHSQVQLVQS GAEVKKPGSS VKVSCKASGN SFSNHDVHWV RQATGQGLEW MGWMSHEGDK TGLAQKFQG RVTITRDSGA STVYMELRGL TADDTAIYYC LTGSKHRLRD YFLYNEYGPN YEEWGDYLAT LDVWGHGTAV T VSSASTKG PSVFPLAPSS KSTSGGTAAL GCLVKDYFPE PVTVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LG TQTYICN VNHKPSNTKV DKKVEPKSCD |
-Macromolecule #4: PGT145 antibody fragment, light chain
| Macromolecule | Name: PGT145 antibody fragment, light chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.95375 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVVITQSPLF LPVTPGEAAS LSCKCSHSLQ HSTGANYLAW YLQRPGQTPR LLIHLATHRA SGVPDRFSGS GSGTDFTLKI SRVESDDVG TYYCMQGLHS PWTFGQGTKV EIKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV ...String: EVVITQSPLF LPVTPGEAAS LSCKCSHSLQ HSTGANYLAW YLQRPGQTPR LLIHLATHRA SGVPDRFSGS GSGTDFTLKI SRVESDDVG TYYCMQGLHS PWTFGQGTKV EIKRTVAAPS VFIFPPSDEQ LKSGTASVVC LLNNFYPREA KVQWKVDNAL Q SGNSQESV TEQDSKDSTY SLSSTLTLSK ADYEKHKVYA CEVTHQGLSS PVTKSFNRGE C |
-Macromolecule #11: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 11 / Number of copies: 20 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 162.24 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.75 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Rigid body fitting followed by real space refinement to improve map occupancy |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-8jtd: |
 Movie
Movie Controller
Controller




 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)