+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a designed AAV8-based vector | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAV8 / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationT=1 icosahedral viral capsid / nucleotide binding / structural molecule activity Similarity search - Function | |||||||||
| Biological species |  Adeno-associated virus - 8 Adeno-associated virus - 8 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.08 Å | |||||||||
 Authors Authors | Ke X / Luo S / Zheng Q / Jiang H / Liu F / Sun X | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: An adeno-associated virus variant enabling efficient ocular-directed gene delivery across species. Authors: Shuang Luo / Hao Jiang / Qingwei Li / Yingfei Qin / Shiping Yang / Jing Li / Lingli Xu / Yan Gou / Yafei Zhang / Fengjiang Liu / Xiao Ke / Qiang Zheng / Xun Sun /  Abstract: Recombinant adeno-associated viruses (rAAVs) have emerged as promising gene therapy vectors due to their proven efficacy and safety in clinical applications. In non-human primates (NHPs), rAAVs are ...Recombinant adeno-associated viruses (rAAVs) have emerged as promising gene therapy vectors due to their proven efficacy and safety in clinical applications. In non-human primates (NHPs), rAAVs are administered via suprachoroidal injection at a higher dose. However, high doses of rAAVs tend to increase additional safety risks. Here, we present a novel AAV capsid (AAVv128), which exhibits significantly enhanced transduction efficiency for photoreceptors and retinal pigment epithelial (RPE) cells, along with a broader distribution across the layers of retinal tissues in different animal models (mice, rabbits, and NHPs) following intraocular injection. Notably, the suprachoroidal delivery of AAVv128-anti-VEGF vector completely suppresses the Grade IV lesions in a laser-induced choroidal neovascularization (CNV) NHP model for neovascular age-related macular degeneration (nAMD). Furthermore, cryo-EM analysis at 2.1 Å resolution reveals that the critical residues of AAVv128 exhibit a more robust advantage in AAV binding, the nuclear uptake and endosome escaping. Collectively, our findings highlight the potential of AAVv128 as a next generation ocular gene therapy vector, particularly using the suprachoroidal delivery route. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36594.map.gz emd_36594.map.gz | 176.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36594-v30.xml emd-36594-v30.xml emd-36594.xml emd-36594.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36594.png emd_36594.png | 56.8 KB | ||
| Filedesc metadata |  emd-36594.cif.gz emd-36594.cif.gz | 5.5 KB | ||
| Others |  emd_36594_half_map_1.map.gz emd_36594_half_map_1.map.gz emd_36594_half_map_2.map.gz emd_36594_half_map_2.map.gz | 318.6 MB 318.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36594 http://ftp.pdbj.org/pub/emdb/structures/EMD-36594 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36594 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36594 | HTTPS FTP |
-Validation report
| Summary document |  emd_36594_validation.pdf.gz emd_36594_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36594_full_validation.pdf.gz emd_36594_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_36594_validation.xml.gz emd_36594_validation.xml.gz | 17.4 KB | Display | |
| Data in CIF |  emd_36594_validation.cif.gz emd_36594_validation.cif.gz | 20.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36594 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36594 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36594 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36594 | HTTPS FTP |
-Related structure data
| Related structure data |  8jreMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36594.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36594.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.73 Å | ||||||||||||||||||||||||||||||||||||
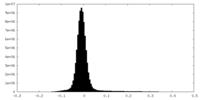
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36594_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
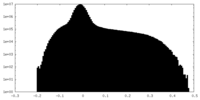
| Density Histograms |
-Half map: #1
| File | emd_36594_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
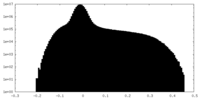
| Density Histograms |
- Sample components
Sample components
-Entire : Adeno-associated virus - 8
| Entire | Name:  Adeno-associated virus - 8 Adeno-associated virus - 8 |
|---|---|
| Components |
|
-Supramolecule #1: Adeno-associated virus - 8
| Supramolecule | Name: Adeno-associated virus - 8 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 202813 / Sci species name: Adeno-associated virus - 8 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 60 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Adeno-associated virus - 8 Adeno-associated virus - 8 |
| Molecular weight | Theoretical: 59.270191 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GADGVGSSSG NWHCDSTWLG DRVITTSTRT WALPTYNNHL YKQISNGTSG GATNDNTYFG YSTPWGYFDF NRFHCHFSPR DWQRLINNN WGFRPKRLSF KLFNIQVKEV TQNEGTKTIA NNLTSTIQVF TDSEYQLPYV LGSAHQGCLP PFPADVFMIP Q YGYLTLNN ...String: GADGVGSSSG NWHCDSTWLG DRVITTSTRT WALPTYNNHL YKQISNGTSG GATNDNTYFG YSTPWGYFDF NRFHCHFSPR DWQRLINNN WGFRPKRLSF KLFNIQVKEV TQNEGTKTIA NNLTSTIQVF TDSEYQLPYV LGSAHQGCLP PFPADVFMIP Q YGYLTLNN GSQAVGRSSF YCLEYFPSQM LRTGNNFQFT YTFEDVPFHS SYAHSQSLDR LMNPLIDQYL YYLSRTQTTG GT ANTQTLG FSQGGPNTMA NQAKNWLPGP CYRQQRVSTT TGQNNNSNFA WTAGTKYHLN GRNSLANPGI AMATHKDDEE RFF PSNGIL IFGKQNAARD NADYSDVMLT SEEEIKTTNP VATEEYGIVA DNLQRGNQQN TARQPQIGTV NSQGALPGMV WQNR DVYLQ GPIWAKIPHT DGNFHPSPLM GGFGLKHPPP QILIKNTPVP ADPPTTFNQS KLNSFITQYS TGQVSVEIEW ELQKE NSKR WNPEIQYTSN YYKSTSVDFA VNTEGVYSEP RPIGTRYLTR NL UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.08 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 90744 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)